Page 104 - Read Online
P. 104
Amatori et al. RT-qPCR array for gene expression
from the RNA extraction procedure, we also designed Thermo Fisher Scientific, 88 genes using real Time
a primer pair that amplified an intronic region of the PCR Assay Panels by BioRad), increasing the risk of
housekeeping gene GAPDH. Importantly, since each not identifying possible technical errors.
sample of cDNA would be amplified in a separated
100-well disc, we also added an additional control to Prior to using the array in an experimental model,
verify the “inter-disc” variability by using, as template, we tested 4 commercially available SYBR Green
a known amount of human genomic DNA and a primer master mixes, focusing attention on 3 genes that can
pair that amplified genomic GAPDH. recapitulate 3 different levels of gene expression: high
levels (GAPDH), medium/low levels (CDKN1A), and
To further increase the reliability of the assay, very low levels (CDKN2B). In fact, it is known that
amplification of each target gene was performed in expression level of some genes involved in the DNA-
duplicate. Most commercially available PCR arrays damage response (e.g. genes that are involved in cell
contain more target genes but lacks replicates (e.g. cycle regulation, such as CDK-inhibitors) are at the
84 genes using the RT Profiler PCR arrays by limit of the detection and thus require a highly efficient
TM
2
Qiagen, 92 genes using TaqMan Array Plates by DNA-polymerase. Therefore, we optimized our RT-
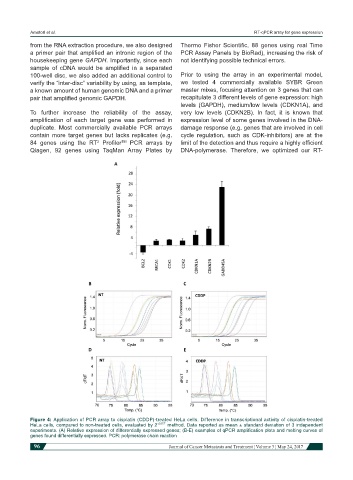
Figure 4: Application of PCR array to cisplatin (CDDP)-treated HeLa cells. Difference in transcriptional activity of cisplatin-treated
HeLa cells, compared to non-treated cells, evaluated by 2 -∆∆CT method. Data reported as mean ± standard deviation of 3 independent
experiments. (A) Relative expression of differentially expressed genes; (B-E) examples of qPCR amplification plots and melting curves of
genes found differentially expressed. PCR: polymerase chain reaction
96 Journal of Cancer Metastasis and Treatment ¦ Volume 3 ¦ May 24, 2017