Page 102 - Read Online
P. 102
Amatori et al. RT-qPCR array for gene expression
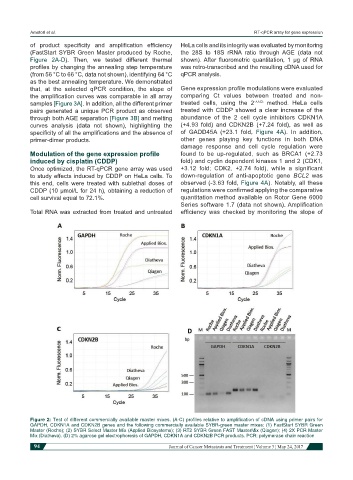
of product specificity and amplification efficiency HeLa cells and its integrity was evaluated by monitoring
(FastStart SYBR Green Master produced by Roche, the 28S to 18S rRNA ratio through AGE (data not
Figure 2A-D). Then, we tested different thermal shown). After fluorometric quantitation, 1 µg of RNA
profiles by changing the annealing step temperature was retro-transcribed and the resulting cDNA used for
(from 56 °C to 66 °C, data not shown), identifying 64 °C qPCR analysis.
as the best annealing temperature. We demonstrated
that, at the selected qPCR condition, the slope of Gene expression profile modulations were evaluated
the amplification curves was comparable in all array comparing Ct values between treated and non-
samples [Figure 3A]. In addition, all the different primer treated cells, using the 2 -∆∆Ct method. HeLa cells
pairs generated a unique PCR product as observed treated with CDDP showed a clear increase of the
through both AGE separation [Figure 3B] and melting abundance of the 2 cell cycle inhibitors CDKN1A
curves analysis (data not shown), highlighting the (+4.93 fold) and CDKN2B (+7.24 fold), as well as
specificity of all the amplifications and the absence of of GADD45A (+23.1 fold, Figure 4A). In addition,
primer-dimer products. other genes playing key functions in both DNA
damage response and cell cycle regulation were
Modulation of the gene expression profile found to be up-regulated, such as BRCA1 (+2.73
induced by cisplatin (CDDP) fold) and cyclin dependent kinases 1 and 2 (CDK1,
Once optimized, the RT-qPCR gene array was used +3.12 fold; CDK2, +2.74 fold), while a significant
to study effects induced by CDDP on HeLa cells. To down-regulation of anti-apoptotic gene BCL2 was
this end, cells were treated with sublethal doses of observed (-3.63 fold, Figure 4A). Notably, all these
CDDP (10 µmol/L for 24 h), obtaining a reduction of regulations were confirmed applying the comparative
cell survival equal to 72.1%. quantitation method available on Rotor Gene 6000
Series software 1.7 (data not shown). Amplification
Total RNA was extracted from treated and untreated efficiency was checked by monitoring the slope of
Figure 2: Test of different commercially available master mixes. (A-C) profiles relative to amplification of cDNA using primer pairs for
GAPDH, CDKN1A and CDKN2B genes and the following commercially available SYBR-green master mixes: (1) FastStart SYBR Green
Master (Roche); (2) SYBR Select Master Mix (Applied Biosystems); (3) RT2 SYBR Green FAST MasterMix (Qiagen); (4) 2X PCR Master
Mix (Diatheva). (D) 2% agarose gel electrophoresis of GAPDH, CDKN1A and CDKN2B PCR products. PCR: polymerase chain reaction
94 Journal of Cancer Metastasis and Treatment ¦ Volume 3 ¦ May 24, 2017