Page 100 - Read Online
P. 100
Amatori et al. RT-qPCR array for gene expression
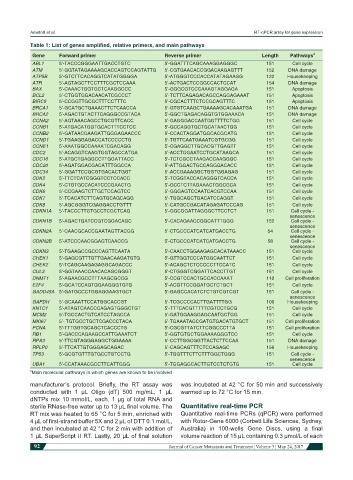
Table 1: List of genes amplified, relative primers, and main pathways
Gene Forward primer Reverse primer Length Pathways a
ABL1 5’-TACCCGGGAATTGACCTGTC 5’-GGATTTCAGCAAAGGAGGGC 151 Cell cycle
ATM 5’-GGTATAGAAAAGCACCAGTCCAGTATTG 5’-CGTGAACACCGGACAAGAGTTT 152 DNA damage
ATP5B 5’-GTCTTCACAGGTCATATGGGGA 5’-ATGGGTCCCACCATATAGAAGG 122 Housekeeping
ATR 5’-AGTAGCTTCCTTTCGCTCCAAA 5’-ACTGACTCCGGCCACTCCAT 154 DNA damage
BAX 5’-CAAACTGGTGCTCAAGGCCC 5’-GGGCGTCCCAAAGTAGGAGA 151 Apoptosis
BCL2 5’-CTGGTGGACAACATCGCCCT 5’-TCTTCAGAGACAGCCAGGAGAAAT 151 Apoptosis
BIRC5 5’-CCGGTTGCGCTTTCCTTTC 5’-CGCACTTTCTCCGCAGTTTC 151 Apoptosis
BRCA1 5’-GCATGCTGAAACTTCTCAACCA 5’-GTGTCAAGCTGAAAAGCACAAATGA 151 DNA damage
BRCA2 5’-AGACTGTACTTCAGGGCCGTACA 5’-GGCTGAGACAGGTGTGGAAACA 151 DNA damage
CCNA2 5’-AGTAAACAGCCTGCGTTCACC 5’-GAGGGACCAATGGTTTTCTGG 151 Cell cycle
CCNB1 5’-ATGACATGGTGCACTTTCCTCC 5’-GCCAGGTGCTGCATAACTGG 151 Cell cycle
CCNB2 5’-GATAACGAAGATTGGGAGAACCC 5’-CCACTAGGATGGCACGCATG 151 Cell cycle
CCND1 5’-TGAAGGAGACCATCCCCCTG 5’-TGTTCAATGAAATCGTGCGG 151 Cell cycle
CCNE1 5’-AAATGGCCAAAATCGACAGG 5’-CGAGGCTTGCACGTTGAGTT 151 Cell cycle
CDC2 5’-ACAGGTCAAGTGGTAGCCATGA 5’-ACCTGGAATCCTGCATAAGCA 151 Cell cycle
CDC16 5’-ATGCTGAGGCCTTGGATTACC 5’-TCTCGCCTAAGACCAAGGGC 151 Cell cycle
CDC20 5’-AGATGGACGACATTTGGCCA 5’-ATTGGACTGCCAGGGACACC 151 Cell cycle
CDC34 5’-GGATTCCGCGTGACACTGGT 5’-ACCGAAAGGCTGGTGGAGAG 151 Cell cycle
CDK2 5’-TTCTCATCGGGTCCTCCACC 5’-TCGGTACCACAGGGTCACCA 151 Cell cycle
CDK4 5’-CTGTGCCACATCCCGAACTG 5’-GCCTCTTAGAAACTGGCGCA 151 Cell cycle
CDK6 5’-CCGAAGTCTTGCTCCAGTCC 5’-GGGAGTCCAATCACGTCCAA 151 Cell cycle
CDK7 5’-TCACATCTTCAGTGCAGCAGG 5’-TGGCAGCTGACATCCAGGT 151 Cell cycle
CDK8 5’-AGCGGGTCGAGGACCTGTTT 5’-CATGCCGACATAGAGATCCCAG 151 Cell cycle
CDKN1A 5’-TACCCTTGTGCCTCGCTCAG 5’-GGCGGATTAGGGCTTCCTCT 151 Cell cycle -
senescence
CDKN1B 5’-AGACTGATCCGTCGGACAGC 5’-CACAGAACCGGCATTTGGG 152 Cell cycle -
senescence
CDKN2A 5’-CAACGCACCGAATAGTTACGG 5’-CTGCCCATCATCATGACCTG 54 Cell cycle -
senescence
CDKN2B 5’-ATCCCAACGGAGTCAACCG 5’-CTGCCCATCATCATGACCTG 58 Cell cycle -
senescence
CDKN3 5’-TGAAGCCGCCCAGTTCAATA 5’-CAACCTGGAAGAGCACATAAACC 151 Cell cycle
CHEK1 5’-GAGCGTTTGTTGAACAAGATGTG 5’-GTTGGTCCCATGGCAATTCT 151 Cell cycle
CHEK2 5’-TCAGCAAGAGAGGCAGACCC 5’-ACAGCTCTCCCCCTTCCATC 151 Cell cycle
CUL3 5’-GGTAAACCAACACAGCGGGT 5’-CTGGGTCGGATTCACCTTGT 151 Cell cycle
DNMT1 5’-AGAACGCCTTTAAGCGCCG 5’-CCGTCCACTGCCACCAAAT 110 Cell proliferation
E2F4 5’-GCATCCAGTGGAAGGGTGTG 5’-ACGTTCCGGATGCTCTGCT 151 Cell cycle
GADD45A 5’-GATGCCCTGGAGGAAGTGCT 5’-GAGCCACATCTCTGTCGTCGT 151 Cell cycle -
senescence
GAPDH 5’-GCAAATTCCATGGCACCGT 5’-TCGCCCCACTTGATTTTGG 106 Housekeeping
KNTC1 5’-ATAGTCAACCCAGAGTGGGCTGT 5’-TTTCACGTTTTTCGTCCTGCG 151 Cell cycle
MCM2 5’-TGCCACTGTCATCCTAGCCA 5’-GATGGAAGGAGCAATGCTGG 151 Cell cycle
MKI67 5’- TGTGCCTGCTCGACCCTACA 5’-TGAAATAGCGATGTGACATGTGCT 151 Cell proliferation
PCNA 5’-TTTGGTGCAGCTCACCCTG 5’-CGCGTTATCTTCGGCCCTTA 151 Cell proliferation
RB1 5’-GACCCAGAAGCCATTGAAATCT 5’-GGTGTGCTGGAAAAGGGTCC 151 Cell cycle
RPA3 5’-TTCGTAGGGAGGCTGGAAAA 5’-CCTTGGCGGTTACTCTTCCAA 151 DNA damage
RPLP0 5’-TTCATTGTGGGAGCAGAC 5’-CAGCAGTTTCTCCAGAGC 156 Housekeeping
TP53 5’-GCGTGTTTGTGCCTGTCCTG 5’-TGGTTTCTTCTTTGGCTGGG 151 Cell cycle -
senescence
UBA1 5’-CCATAAACGCCTTCATTGGG 5’-TGGAGGCACTTGTCCTCTGTG 151 Cell cycle
a Main molecular pathways in which genes are known to be involved
manufacturer’s protocol. Briefly, the RT assay was was incubated at 42 °C for 50 min and successively
conducted with 1 µL Oligo (dT) 500 mg/mL, 1 µL warmed up to 72 °C for 15 min.
dNTPs mix 10 mmol/L, each, 1 µg of total RNA and
sterile RNase-free water up to 13 µL final volume. The Quantitative real-time PCR
RT mix was heated to 65 °C for 5 min, enriched with Quantitative real-time PCRs (qPCR) were performed
4 µL of first-strand buffer 5X and 2 µL of DTT 0.1 mol/L, with Rotor-Gene 6000 (Corbett Life Sciences, Sydney,
and then incubated at 42 °C for 2 min with addition of Australia) in 100-wells Gene Discs, using a final
1 µL SuperScript II RT. Lastly, 20 µL of final solution volume reaction of 15 µL containing 0.3 µmol/L of each
92 Journal of Cancer Metastasis and Treatment ¦ Volume 3 ¦ May 24, 2017