Page 165 - Read Online
P. 165
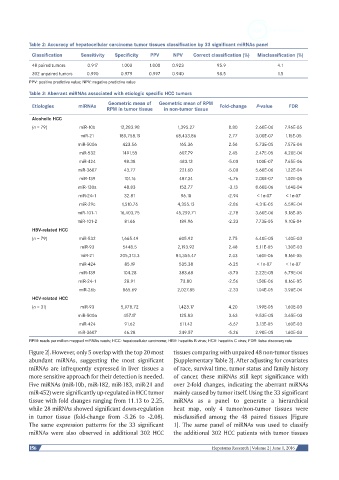
Table 2: Accuracy of hepatocellular carcinoma tumor tissues classification by 33 significant miRNAs panel
Classification Sensitivity Specificity PPV NPV Correct classification (%) Misclassification (%)
48 paired tumors 0.917 1.000 1.000 0.923 95.9 4.1
302 unpaired tumors 0.990 0.979 0.997 0.940 98.5 1.5
PPV: positive predictive value; NPV: negative predictive value
Table 3: Aberrant miRNAs associated with etiologic specific HCC tumors
Geometric mean of Geometric mean of RPM
Etiologies miRNAs Fold-change P-value FDR
RPM in tumor tissue in non-tumor tissue
Alcoholic HCC
(n = 79) miR-10b 12,283.98 1,395.27 8.80 2.60E-06 7.96E-05
miR-21 189,758.19 68,433.86 2.77 3.00E-07 1.15E-05
miR-500a 423.56 165.36 2.56 5.73E-05 7.57E-04
miR-532 1491.55 607.79 2.45 2.47E-05 4.20E-04
miR-424 98.38 483.13 -5.00 1.00E-07 7.65E-06
miR-3607 43.77 221.60 -5.00 5.60E-06 1.22E-04
miR-139 101.16 487.34 -4.76 2.00E-07 1.02E-05
miR-130a 48.83 152.77 -3.13 8.60E-06 1.64E-04
miR-24-1 32.81 96.18 -2.94 < 1e-07 < 1e-07
miR-29c 1,510.76 4,355.13 -2.86 4.31E-05 6.59E-04
miR-101-1 16,403.75 45,299.71 -2.78 3.60E-06 9.18E-05
miR-101-2 81.66 189.96 -2.33 7.73E-05 9.10E-04
HBV-related HCC
(n = 79) miR-532 1,665.49 605.92 2.75 6.40E-05 1.40E-03
miR-93 5448.5 2,193.92 2.48 5.11E-05 1.30E-03
miR-21 205,313.3 84,355.47 2.43 1.60E-06 8.16E-05
miR-424 85.19 535.38 -6.25 < 1e-07 < 1e-07
miR-139 104.28 383.68 -3.70 2.22E-05 6.79E-04
miR-24-1 28.91 73.80 -2.56 1.50E-06 8.16E-05
miR-26b 865.69 2,027.85 -2.33 1.04E-05 3.98E-04
HCV-related HCC
(n = 31) miR-93 5,978.72 1,423.17 4.20 1.99E-05 1.60E-03
miR-500a 457.17 125.83 3.63 9.53E-05 3.65E-03
miR-424 91.62 611.42 -6.67 3.13E-05 1.60E-03
miR-3607 46.28 249.57 -5.26 2.90E-05 1.60E-03
RPM: reads per million mapped miRNAs reads; HCC: hepatocellular carcinoma; HBV: hepatitis B virus; HCV: hepatitis C virus; FDR: false discovery rate
Figure 2]. However, only 5 overlap with the top 20 most tissues comparing with unpaired 48 non-tumor tissues
abundant miRNAs, suggesting the most significant [Supplementary Table 2]. After adjusting for covariates
miRNAs are infrequently expressed in liver tissues a of race, survival time, tumor status and family history
more sensitive approach for their detection is needed. of cancer, these miRNAs still kept significance with
Five miRNAs (miR-10b, miR-182, miR-183, miR-21 and over 2-fold changes, indicating the aberrant miRNAs
miR-452) were significantly up-regulated in HCC tumor mainly caused by tumor itself. Using the 33 significant
tissue with fold changes ranging from 11.13 to 2.25, miRNAs as a panel to generate a hierarchical
while 28 miRNAs showed significant down-regulation heat map, only 4 tumor/non-tumor tissues were
in tumor tissue (fold-change from -5.26 to -2.08). misclassified among the 48 paired tissues [Figure
The same expression patterns for the 33 significant 1]. The same panel of miRNAs was used to classify
miRNAs were also observed in additional 302 HCC the additional 302 HCC patients with tumor tissues
156 Hepatoma Research | Volume 2 | June 1, 2016