Page 164 - Read Online
P. 164
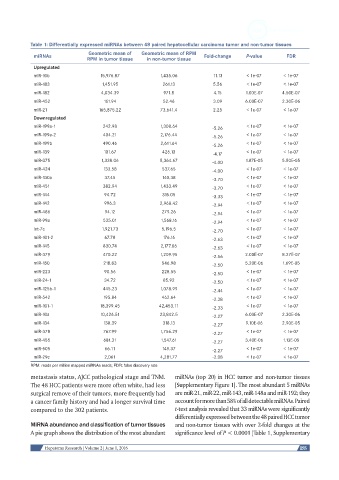
Table 1: Differentially expressed miRNAs between 48 paired hepatocellular carcinoma tumor and non-tumor tissues
Geometric mean of Geometric mean of RPM
miRNAs Fold-change P-value FDR
RPM in tumor tissue in non-tumor tissue
Upregulated
miR-10b 15,976.87 1,435.06 11.13 < 1e-07 < 1e-07
miR-183 1,451.95 261.13 5.56 < 1e-07 < 1e-07
miR-182 4,034.39 971.8 4.15 1.00E-07 4.50E-07
miR-452 161.94 52.46 3.09 6.00E-07 2.30E-06
miR-21 165,875.22 73,641.4 2.25 < 1e-07 < 1e-07
Downregulated
miR-199a-1 242.98 1,308.64 -5.26 < 1e-07 < 1e-07
miR-199a-2 404.21 2,176.44 -5.26 < 1e-07 < 1e-07
miR-199b 490.46 2,611.64 -5.26 < 1e-07 < 1e-07
miR-139 101.67 426.13 -4.17 < 1e-07 < 1e-07
miR-375 1,338.06 5,364.67 -4.00 1.87E-05 5.50E-05
miR-424 133.58 537.65 -4.00 < 1e-07 < 1e-07
miR-130a 37.45 140.38 -3.70 < 1e-07 < 1e-07
miR-451 382.94 1,433.49 -3.70 < 1e-07 < 1e-07
miR-144 94.72 315.05 -3.33 < 1e-07 < 1e-07
miR-142 996.3 2,968.42 -2.94 < 1e-07 < 1e-07
miR-486 94.12 279.26 -2.94 < 1e-07 < 1e-07
miR-99a 535.01 1,568.16 -2.94 < 1e-07 < 1e-07
let-7c 1,921.73 5,196.5 -2.70 < 1e-07 < 1e-07
miR-101-2 67.78 176.16 -2.63 < 1e-07 < 1e-07
miR-145 830.74 2,177.06 -2.63 < 1e-07 < 1e-07
miR-379 470.22 1,209.95 -2.56 2.00E-07 8.27E-07
miR-150 218.83 546.98 -2.50 5.20E-06 1.69E-05
miR-223 90.56 228.55 -2.50 < 1e-07 < 1e-07
miR-24-1 34.72 85.92 -2.50 < 1e-07 < 1e-07
miR-125b-1 445.23 1,078.99 -2.44 < 1e-07 < 1e-07
miR-542 195.84 462.64 -2.38 < 1e-07 < 1e-07
miR-101-1 18,399.45 42,453.11 -2.33 < 1e-07 < 1e-07
miR-10a 10,426.51 23,842.5 -2.27 6.00E-07 2.30E-06
miR-134 138.39 318.13 -2.27 9.10E-06 2.90E-05
miR-378 767.99 1,756.29 -2.27 < 1e-07 < 1e-07
miR-455 681.31 1,547.61 -2.27 3.40E-06 1.13E-05
miR-505 66.11 149.37 -2.27 < 1e-07 < 1e-07
miR-29c 2,061 4,281.77 -2.08 < 1e-07 < 1e-07
RPM: reads per million mapped miRNAs reads; FDR: false discovery rate
metastasis status, AJCC pathological stage and TNM. miRNAs (top 20) in HCC tumor and non-tumor tissues
The 48 HCC patients were more often white, had less [Supplementary Figure 1]. The most abundant 5 miRNAs
surgical remove of their tumors, more frequently had are miR-21, miR-22, miR-143, miR-148a and miR-192; they
a cancer family history and had a longer survival time account for more than 58% of all detectable miRNAs. Paired
compared to the 302 patients. t-test analysis revealed that 33 miRNAs were significantly
differentially expressed between the 48 paired HCC tumor
MiRNA abundance and classification of tumor tissues and non-tumor tissues with over 2-fold changes at the
A pie graph shows the distribution of the most abundant significance level of P < 0.0001 [Table 1, Supplementary
Hepatoma Research | Volume 2 | June 1, 2016 155