Page 245 - Read Online
P. 245
Raevsky et al. Neuroimmunol Neuroinflammation 2018;5:33 I http://dx.doi.org/10.20517/2347-8659.2018.34 Page 7 of 10
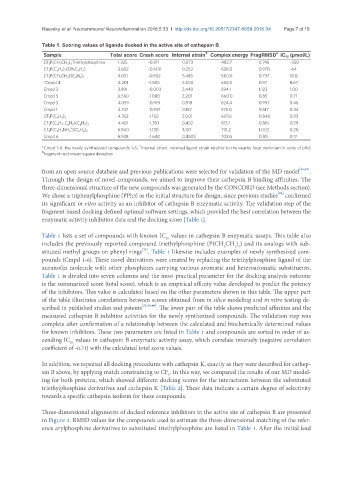
Table 1. Scoring values of ligands docked in the active site of cathepsin B
₸
Sample Total score Crash score Internal strain Complex energy FragRMSD IC 50 (µmol/L)
#
CF 3 P(CH 2 CH 3 ) 3 Triethylphosphine 1.325 -0.211 0.073 482.7 0.745 ~250
CF 3 P(C 6 H 5 ) 2 (CH 2 C 6 H 5 ) 3.602 -0.600 0.252 626.0 0.970 ~64
4.051 -0.952 5.485 560.0 0.737 18.0
CF 3 P(CH 2 CH 3 )(C 6 H 5 ) 2
*Cmpd 4 4.201 -1.505 2.500 658.5 0.97 8.67
Cmpd 2 3.991 -0.003 2.449 594.1 1.123 1.00
Cmpd 5 6.560 -1.080 2.201 660.0 0.85 0.71
Cmpd 3 4.059 -0.919 0.918 624.4 0.997 0.46
Cmpd 1 4.747 -0.937 0.157 575.0 0.617 0.34
4.353 -1.150 2.001 607.6 0.840 0.33
CF 3 P(C 6 H 5 ) 3
4.401 -1.301 0.452 613.1 0.861 0.29
CF 3 P(C 6 H 4 -C 6 H 5 )(C 6 H 5 ) 2
+
CF 3 P(C 6 H 4 NH 3 )(C 6 H 5 ) 2 6.940 -1.130 3.157 731.2 1.002 0.20
Cmpd 6 6.938 -1.640 0.4503 703.6 0.80 0.17
₸
*Cmpd 1-6: the newly synthesized compounds 1-6; Internal strain: nominal ligand strain relative to the nearby local minimum in units of pKd;
# fragment root-mean-square deviation
from an open source database and previous publications were selected for validation of the MD model [36,39] .
Through the design of novel compounds, we aimed to improve their cathepsin B binding affinities. The
three-dimensional structure of the new compounds was generated by the CONCORD (see Methods section).
[29]
We chose a triphenylphosphine (PPh3) as the initial structure for design, since previous studies confirmed
its significant in vitro activity as an inhibitor of cathepsin B enzymatic activity. The validation step of the
fragment-based docking defined optimal software settings, which provided the best correlation between the
enzymatic activity inhibition data and the docking score [Table 1].
Table 1 lists a set of compounds with known IC values in cathepsin B enzymatic assays. This table also
50
includes the previously reported compound triethylphosphine (P(CH CH ) ) and its analogs with sub-
3 3
2
[31]
stituted methyl groups on phenyl rings . Table 1 likewise includes examples of newly synthesized com-
pounds (Cmpd 1-6). These novel derivatives were created by replacing the triethylphosphine ligand of the
auranofin molecule with other phosphines carrying various aromatic and heteroaromatic substituents.
Table 1 is divided into seven columns and the most practical parameter for the docking analysis outcome
is the summarized score (total score), which is an empirical affinity value developed to predict the potency
of the inhibitors. This value is calculated based on the other parameters shown in this table. The upper part
of the table illustrates correlations between scores obtained from in silico modeling and in vitro testing de-
scribed in published studies and patents [31,36,40] . The lower part of the table shows predicted affinities and the
measured cathepsin B inhibitor activities for the newly synthesized compounds. The validation step was
complete after confirmation of a relationship between the calculated and biochemically determined values
for known inhibitors. These two parameters are listed in Table 1 and compounds are sorted in order of as-
cending IC values in cathepsin B enzymatic activity assay, which correlate inversely (negative correlation
50
coefficient of -0.71) with the calculated total score values.
In addition, we repeated all docking procedures with cathepsin K, exactly as they were described for cathep-
sin B above, by applying match constraining to CF . In this way, we compared the results of our MD model-
3
ing for both proteins, which showed different docking scores for the interactions between the substituted
triethylphosphine derivatives and cathepsin K [Table 2]. These data indicate a certain degree of selectivity
towards a specific cathepsin isoform for these compounds.
Three-dimensional alignments of docked reference inhibitors in the active site of cathepsin B are presented
in Figure 3. RMSD values for the compounds used to estimate the three-dimensional matching of the refer-
ence arylphosphine derivatives to substituted triethylphosphine are listed in Table 1. After the initial lead