Page 83 - Read Online
P. 83
Page 152 Goodman et al. J Transl Genet Genom 2020;4:144-58 I http://dx.doi.org/10.20517/jtgg.2020.23
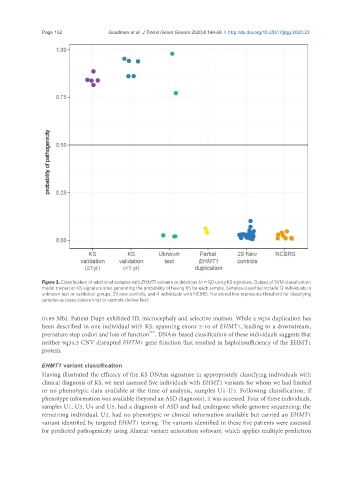
Figure 3. Classification of additional samples with EHMT1 variants or deletions (n = 12) using KS signature. Output of SVM classification
model trained on KS signature sites generating the probability of having KS for each sample. Samples classified include 12 individuals in
unknown test or validation groups, 29 new controls, and 4 individuals with NCBRS. Horizontal line represents threshold for classifying
samples as cases (above line) or controls (below line)
(0.89 Mb). Patient Dup2 exhibited ID, microcephaly and selective mutism. While a 9q34 duplication has
been described in one individual with KS, spanning exons 2-10 of EHMT1, leading to a downstream,
premature stop codon and loss of function . DNAm-based classification of these individuals suggests that
[37]
neither 9q34.3 CNV disrupted EHTM1 gene function that resulted in haploinsufficiency of the EHMT1
protein.
EHMT1 variant classification
Having illustrated the efficacy of the KS DNAm signature in appropriately classifying individuals with
clinical diagnosis of KS, we next assessed five individuals with EHMT1 variants for whom we had limited
or no phenotypic data available at the time of analysis, samples U1-U5. Following classification, if
phenotype information was available (beyond an ASD diagnosis), it was accessed. Four of these individuals,
samples U1, U3, U4 and U5, had a diagnosis of ASD and had undergone whole-genome sequencing; the
remaining individual, U2, had no phenotypic or clinical information available but carried an EHMT1
variant identified by targeted EHMT1 testing. The variants identified in these five patients were assessed
for predicted pathogenicity using Alamut variant annotation software, which applies multiple prediction