Page 82 - Read Online
P. 82
Goodman et al. J Transl Genet Genom 2020;4:144-58 I http://dx.doi.org/10.20517/jtgg.2020.23 Page 151
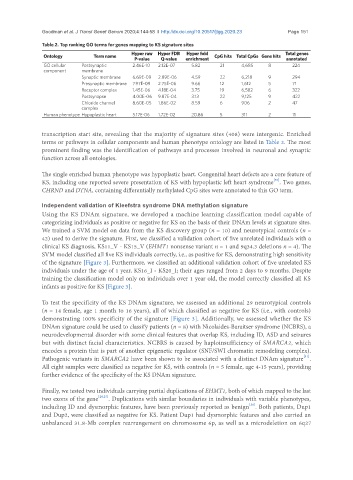
Table 2. Top ranking GO terms for genes mapping to KS signature sites
Hyper raw Hyper FDR Hyper fold Total genes
Ontology Term name CpG hits Total CpGs Gene hits
P-value Q-value enrichment annotated
GO cellular Postsynaptic 2.46E-10 2.12E-07 5.82 21 4,685 8 224
component membrane
Synaptic membrane 6.69E-09 2.89E-06 4.59 22 6,218 9 294
Presynaptic membrane 7.97E-09 2.75E-06 9.66 12 1,612 5 71
Receptor complex 1.45E-06 4.18E-04 3.75 19 6,582 6 322
Postsynapse 4.00E-06 9.87E-04 3.13 22 9,125 9 422
Chloride channel 8.60E-05 1.86E-02 8.59 6 906 2 47
complex
Human phenotype Hypoplastic heart 5.17E-06 1.72E-02 20.86 5 311 2 11
transcription start site, revealing that the majority of signature sites (408) were intergenic. Enriched
terms or pathways in cellular components and human phenotype ontology are listed in Table 2. The most
prominent finding was the identification of pathways and processes involved in neuronal and synaptic
function across all ontologies.
The single enriched human phenotype was hypoplastic heart. Congenital heart defects are a core feature of
[35]
KS, including one reported severe presentation of KS with hypoplastic left heart syndrome . Two genes,
CHRND and DTNA, containing differentially methylated CpG sites were annotated to this GO term.
Independent validation of Kleefstra syndrome DNA methylation signature
Using the KS DNAm signature, we developed a machine learning classification model capable of
categorizing individuals as positive or negative for KS on the basis of their DNAm levels at signature sites.
We trained a SVM model on data from the KS discovery group (n = 10) and neurotypical controls (n =
42) used to derive the signature. First, we classified a validation cohort of five unrelated individuals with a
clinical KS diagnosis, KS11_V - KS15_V (EHMT1 nonsense variant n = 1 and 9q34.3 deletions n = 4). The
SVM model classified all five KS individuals correctly, i.e., as positive for KS, demonstrating high sensitivity
of the signature [Figure 3]. Furthermore, we classified an additional validation cohort of five unrelated KS
individuals under the age of 1 year, KS16_I - KS20_I; their ages ranged from 2 days to 9 months. Despite
training the classification model only on individuals over 1 year old, the model correctly classified all KS
infants as positive for KS [Figure 3].
To test the specificity of the KS DNAm signature, we assessed an additional 29 neurotypical controls
(n = 14 female, age 1 month to 16 years), all of which classified as negative for KS (i.e., with controls)
demonstrating 100% specificity of the signature [Figure 3]. Additionally, we assessed whether the KS
DNAm signature could be used to classify patients (n = 8) with Nicolaides-Baraitser syndrome (NCBRS), a
neurodevelopmental disorder with some clinical features that overlap KS, including ID, ASD and seizures
but with distinct facial characteristics. NCBRS is caused by haploinsufficiency of SMARCA2, which
encodes a protein that is part of another epigenetic regulator (SNF/SWI chromatin remodeling complex).
[11]
Pathogenic variants in SMARCA2 have been shown to be associated with a distinct DNAm signature .
All eight samples were classified as negative for KS, with controls (n = 5 female, age 4-15 years), providing
further evidence of the specificity of the KS DNAm signature.
Finally, we tested two individuals carrying partial duplications of EHMT1, both of which mapped to the last
two exons of the gene [26,27] . Duplications with similar boundaries in individuals with variable phenotypes,
[36]
including ID and dysmorphic features, have been previously reported as benign . Both patients, Dup1
and Dup2, were classified as negative for KS. Patient Dup1 had dysmorphic features and also carried an
unbalanced 31.8-Mb complex rearrangement on chromosome 6p, as well as a microdeletion on 6q27