Page 85 - Read Online
P. 85
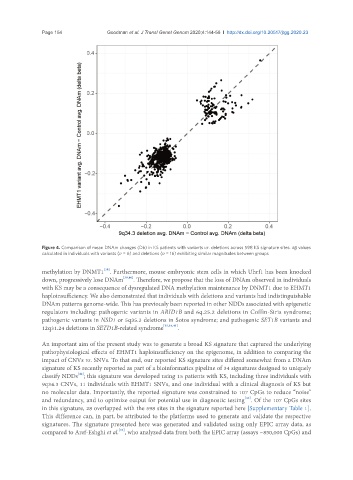
Page 154 Goodman et al. J Transl Genet Genom 2020;4:144-58 I http://dx.doi.org/10.20517/jtgg.2020.23
Figure 4. Comparison of mean DNAm changes (Db) in KS patients with variants vs. deletions across 598 KS signature sites. Δb values
calculated in individuals with variants (n = 6) and deletions (n = 16) exhibiting similar magnitudes between groups
[38]
methylation by DNMT1 . Furthermore, mouse embryonic stem cells in which Uhrf1 has been knocked
down, progressively lose DNAm [39,40] . Therefore, we propose that the loss of DNAm observed in individuals
with KS may be a consequence of dysregulated DNA methylation maintenance by DNMT1 due to EHMT1
haploinsufficiency. We also demonstrated that individuals with deletions and variants had indistinguishable
DNAm patterns genome-wide. This has previously been reported in other NDDs associated with epigenetic
regulators including: pathogenic variants in ARID1B and 6q.25.2 deletions in Coffin-Siris syndrome;
pathogenic variants in NSD1 or 5q35.3 deletions in Sotos syndrome; and pathogenic SET1B variants and
12q31.24 deletions in SETD1B-related syndrome [15,16,41] .
An important aim of the present study was to generate a broad KS signature that captured the underlying
pathophysiological effects of EHMT1 haploinsufficiency on the epigenome, in addition to comparing the
impact of CNVs vs. SNVs. To that end, our reported KS signature sites differed somewhat from a DNAm
signature of KS recently reported as part of a bioinformatics pipeline of 34 signatures designed to uniquely
[42]
classify NDDs ; this signature was developed using 15 patients with KS, including three individuals with
9q34.3 CNVs, 11 individuals with EHMT1 SNVs, and one individual with a clinical diagnosis of KS but
no molecular data. Importantly, the reported signature was constrained to 107 CpGs to reduce “noise”
[42]
and redundancy, and to optimize output for potential use in diagnostic testing . Of the 107 CpGs sites
in this signature, 28 overlapped with the 598 sites in the signature reported here [Supplementary Table 1].
This difference can, in part, be attributed to the platforms used to generate and validate the respective
signatures. The signature presented here was generated and validated using only EPIC array data, as
[42]
compared to Aref-Eshghi et al. , who analyzed data from both the EPIC array (assays ~850,000 CpGs) and