Page 321 - Read Online
P. 321
Shen et al. Evaluation of microRNAs normalization approaches
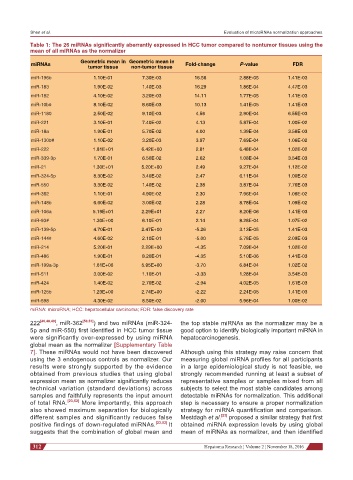
Table 1: The 26 miRNAs significantly aberrantly expressed in HCC tumor compared to nontumor tissues using the
mean of all miRNAs as the normalizer
Geometric mean in Geometric mean in
miRNAs tumor tissue non-tumor tissue Fold-change P-value FDR
miR-196b 1.10E-01 7.30E-03 16.56 2.88E-05 1.41E-03
miR-183 1.90E-02 1.40E-03 16.29 1.86E-04 4.47E-03
miR-182 4.10E-02 3.20E-03 14.11 1.77E-05 1.41E-03
miR-10b# 8.10E-02 8.60E-03 10.13 1.41E-05 1.41E-03
miR-1180 2.50E-02 9.10E-03 4.56 2.90E-04 6.55E-03
miR-221 3.10E-01 7.40E-02 4.13 5.87E-04 1.00E-02
miR-18a 1.90E-01 5.70E-02 4.00 1.39E-04 3.58E-03
miR-130b# 1.10E-02 3.20E-03 3.97 7.69E-04 1.06E-02
miR-222 1.81E+01 6.42E+00 2.81 6.48E-04 1.02E-02
miR-339-3p 1.70E-01 6.50E-02 2.62 1.08E-04 3.54E-03
miR-21 1.30E+01 5.20E+00 2.49 9.27E-04 1.12E-02
miR-324-5p 8.30E-02 3.40E-02 2.47 6.11E-04 1.00E-02
miR-550 3.30E-02 1.40E-02 2.38 3.87E-04 7.76E-03
miR-362 1.10E-01 4.90E-02 2.30 7.96E-04 1.06E-02
miR-148b 6.60E-02 3.00E-02 2.28 8.78E-04 1.09E-02
miR-106a 5.19E+01 2.29E+01 2.27 8.20E-06 1.41E-03
miR-93# 1.30E+00 6.10E-01 2.14 8.28E-04 1.07E-02
miR-139-5p 4.70E-01 2.47E+00 -5.26 3.13E-05 1.41E-03
miR-144# 4.60E-02 2.10E-01 -5.00 5.78E-05 2.09E-03
miR-214 5.20E-01 2.29E+00 -4.35 7.09E-04 1.02E-02
miR-486 1.90E-01 8.20E-01 -4.35 5.10E-06 1.41E-03
miR-199a-3p 1.61E+00 5.95E+00 -3.70 6.84E-04 1.02E-02
miR-511 3.00E-02 1.10E-01 -3.33 1.28E-04 3.54E-03
miR-424 1.40E-02 2.70E-02 -2.94 4.02E-05 1.61E-03
miR-125b 1.23E+00 2.74E+00 -2.22 2.24E-05 1.41E-03
miR-598 4.30E-02 8.50E-02 -2.00 5.96E-04 1.00E-02
miRNA: microRNA; HCC: hepatocellular carcinoma; FDR: false discovery rate
222 [46,48,49] , miR-362 [50,51] ) and two miRNAs (miR-324- the top stable miRNAs as the normalizer may be a
5p and miR-550) first identified in HCC tumor tissue good option to identify biologically important miRNA in
were significantly over-expressed by using miRNA hepatocarcinogenesis.
global mean as the normalizer [Supplementary Table
7]. These miRNAs would not have been discovered Although using this strategy may raise concern that
using the 3 endogenous controls as normalizer. Our measuring global miRNA profiles for all participants
results were strongly supported by the evidence in a large epidemiological study is not feasible, we
obtained from previous studies that using global strongly recommended running at least a subset of
expression mean as normalizer significantly reduces representative samples or samples mixed from all
technical variation (standard deviations) across subjects to select the most stable candidates among
samples and faithfully represents the input amount detectable miRNAs for normalization. This additional
of total RNA. [23,52] More importantly, this approach step is necessary to ensure a proper normalization
also showed maximum separation for biologically strategy for miRNA quantification and comparison.
different samples and significantly reduces false Mestdagh et al. [23] proposed a similar strategy that first
positive findings of down-regulated miRNAs. [23,52] It obtained miRNA expression levels by using global
suggests that the combination of global mean and mean of miRNAs as normalizer, and then identified
312 Hepatoma Research ¦ Volume 2 ¦ November 18, 2016