Page 131 - Read Online
P. 131
Tai et al. Hepatoma Res 2020;6:74 I http://dx.doi.org/10.20517/2394-5079.2020.54 Page 9 of 12
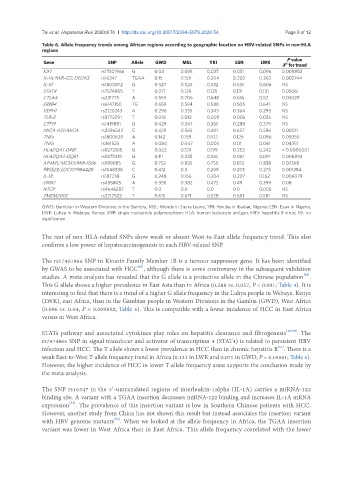
Table 6. Allele frequency trends among African regions according to geographic location on HBV-related SNPs in non-HLA
regions
P value
Gene SNP Allele GWD MSL YRI ESN LWK 2
X for trend
KIF1 rs17401966 G 0.04 0.035 0.037 0.051 0.096 0.009852
IL-1A/MIR-122; DELINS rs16347 TGAA 0.15 0.159 0.204 0.202 0.263 0.002744
IL-10 rs1800872 G 0.527 0.524 0.532 0.551 0.606 NS
STAT4 rs7574865 T 0.071 0.129 0.125 0.131 0.131 0.05661
CTLA4 rs231775 A 0.593 0.706 0.648 0.616 0.52 0.05029
ERBB4 rs6147150 TG 0.659 0.594 0.588 0.505 0.641 NS
VEPH1 rs2120243 A 0.296 0.335 0.343 0.364 0.293 NS
TLR-3 rs3775291 T 0.018 0.012 0.009 0.005 0.035 NS
CTF19 rs1419881 G 0.429 0.347 0.361 0.288 0.379 NS
MICA-AS1/MICA rs2596542 C 0.429 0.565 0.491 0.657 0.586 0.00011
TNFa rs1800629 A 0.142 0.159 0.102 0.126 0.086 0.05056
TNFa rs361525 A 0.084 0.047 0.005 0.01 0.061 0.04351
HLADQA1-DRB1 rs9272105 G 0.062 0.129 0.199 0.253 0.242 < 0.0000001
HLADQA2-DQB1 rs9275319 G 0.111 0.235 0.106 0.061 0.091 0.008894
AP4M1/MCM7/MIR-106b rs999885 G 0.752 0.835 0.755 0.813 0.838 0.07368
PRSS23/LOC107984428 rs1048338 C 0.412 0.3 0.259 0.273 0.273 0.001384
IL-18 rs187238 G 0.248 0.166 0.204 0.207 0.162 0.006079
GRIK1 rs455805 A 0.358 0.382 0.472 0.49 0.399 0.08
NTCP rs4646287 T 0.0 0.0 0.0 0.0 0.005 NS
TMEM200C rs2212522 T 0.615 0.671 0.528 0.581 0.581 NS
GWD: Gambian in Western Divisions in the Gambia; MSL: Mende in Sierra Leone; YRI: Yoruba in Ibadan, Nigeria; ESN: Esan in Nigeria;
LWK: Luhya in Webuye, Kenya; SNP: single nucleotide polymorphism; HLA: human leukocyte antigen; HBV: hepatitis B virus; NS: no
significance
The rest of non-HLA-related SNPs show weak or absent West-to-East allele frequency trend. This also
confirms a low power of hepatocarcinogenesis in each HBV-related SNP.
The rs17401966 SNP in Kinesin Family Member 1B is a tumour suppressor gene. It has been identified
[47]
by GWAS to be associated with HCC , although there is some controversy in the subsequent validation
studies. A meta-analysis has revealed that the G allele is a protective allele in the Chinese population .
[48]
This G allele shows a higher prevalence in East Asia than in Africa (0.288 vs. 0.057, P < 0.001; Table 4). It is
interesting to find that there is a trend of a higher G allele frequency in the Luhya people in Webuye, Kenya
(LWK), east Africa, than in the Gambian people in Western Divisions in the Gambia (GWD), West Africa
(0.096 vs. 0.04, P = 0.009852; Table 6). This is compatible with a lower incidence of HCC in East Africa
versus in West Africa.
STATs pathway and associated cytokines play roles on hepatitis clearance and fibrogenesis [49,50] . The
rs7574865 SNP in signal transducer and activator of transcription 4 (STAT4) is related to persistent HBV
[51]
infection and HCC. The T allele shows a lower prevalence in HCC than in chronic hepatitis B . There is a
weak East-to-West T-allele frequency trend in Africa (0.131 in LWK and 0.071 in GWD; P = 0.05561; Table 6).
However, the higher incidence of HCC in lower T-allele frequency areas supports the conclusion made by
the meta-analysis.
The SNP rs16347 in the 3’-untranslated regions of interleukin-1alpha (IL-1A) carries a miRNA-122
binding site. A variant with a TGAA insertion decreases miRNA-122 binding and increases IL-1A mRNA
expression . The prevalence of this insertion variant is low in Southern Chinese patients with HCC.
[52]
However, another study from China has not shown this result but instead associates the insertion variant
with HBV genome mutants . When we looked at the allele frequency in Africa, the TGAA insertion
[53]
variant was lower in West Africa than in East Africa. This allele frequency correlated with the lower