Page 127 - Read Online
P. 127
Tai et al. Hepatoma Res 2020;6:74 I http://dx.doi.org/10.20517/2394-5079.2020.54 Page 5 of 12
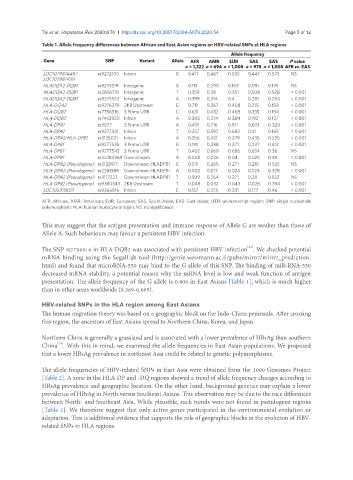
Table 1. Allele frequency differences between African and East Asian regions on HBV-related SNPs at HLA regions
Allele frequency
Gene SNP Variant Allele AFR AMR EUR SAS EAS P value
n = 1,322 n = 694 n = 1,008 n = 978 n = 1,008 AFR vs. EAS
LOC107987449/ rs9272105 Intron G 0.417 0.467 0.535 0.447 0.575 NS
LOC107987459
HLADQA2-DQB1 rs9275319 Intergenic G 0.113 0.290 0.163 0.091 0.135 NS
HLADQA2-DQB1 rs2856718 Intergenic T 0.359 0.38 0.351 0.508 0.528 < 0.001
HLADQA2-DQB1 rs9275572 Intergenic A 0.399 0.314 0.4 0.281 0.254 < 0.001
HLA-DQA2 rs9276370 2KB Upstream G 0.711 0.367 0.408 0.215 0.159 < 0.001
HLA-DQB2 rs7756516 3 Prime UTR C 0.631 0.432 0.468 0.335 0.194 < 0.001
HLA-DQB2 rs7453920 Intron A 0.305 0.274 0.384 0.192 0.127 < 0.001
HLA-DPA1 rs3077 3 Prime UTR A 0.419 0.716 0.811 0.633 0.320 < 0.001
HLA-DPA1 rs9277341 Intron T 0.257 0.597 0.682 0.41 0.165 < 0.001
HLA-DPA1/HLA-DPB1 rs3135021 Intron A 0.356 0.421 0.279 0.435 0.255 < 0.001
HLA-DPB1 rs9277535 3 Prime UTR G 0.191 0.288 0.271 0.297 0.612 < 0.001
HLA-DPB1 rs9277542 3 Prime UTR T 0.402 0.669 0.686 0.634 0.38 NS
HLA-DPB1 rs10484569 Downstream A 0.048 0.036 0.04 0.025 0.39 < 0.001
HLA-DPA2 (Pseudogene) rs3128917 Downstream HLADPB1 G 0.511 0.265 0.271 0.281 0.535 NS
HLA-DPA2 (Pseudogene) rs2281388 Downstream HLADPB1 A 0.002 0.017 0.024 0.024 0.378 < 0.001
HLA-DPA2 (Pseudogene) rs3117222 Downstream HLADPB1 T 0.509 0.264 0.271 0.28 0.537 NS
HLA-DPB2 (Pseudogene) rs9380343 2KB Upstream T 0.048 0.032 0.043 0.028 0.394 < 0.001
LOC105375021 rs9366816 Intron C 0.157 0.375 0.231 0.177 0.46 < 0.001
AFR: African; AMR: American; EUR: European; SAS: South Asian; EAS: East Asian; UTR: un-transcript region; SNP: single nucleotide
polymorphism; HLA: human leukocyte antigen; NS: no significance
This may suggest that the antigen presentation and immune response of Allele G are weaker than those of
Allele A. Such behaviours may favour a persistent HBV infection.
The SNP rs7756516 in HLA-DQB2 was associated with persistent HBV infection . We checked potential
[23]
mRNA binding using the SegalLab tool (http://genie.weizmann.ac.il/pubs/mir07/mir07_prediction.
html) and found that microRNA-550 may bind to the G allele of this SNP. The binding of miR-RNA-550
decreased mRNA stability, a potential reason why the mRNA level is low and weak function of antigen
presentation. The allele frequency of the G allele is 0.806 in East Asians [Table 1], which is much higher
than in other areas worldwide (0.369-0.665).
HBV-related SNPs in the HLA region among East Asians
The human migration theory was based on a geographic block on the Indo-China peninsula. After crossing
this region, the ancestors of East Asians spread to Northern China, Korea, and Japan.
Northern China is generally a grassland and is associated with a lower prevalence of HBsAg than southern
[16]
China . With this in mind, we examined the allele frequencies in East Asian populations. We proposed
that a lower HBsAg prevalence in northeast Asia could be related to genetic polymorphisms.
The allele frequencies of HBV-related SNPs in East Asia were obtained from the 1000 Genomes Project
[Table 2]. A zone in the HLA-DP and -DQ regions showed a trend of allele frequency changes according to
HBsAg prevalence and geographic location. On the other hand, background genetics may explain a lower
prevalence of HBsAg in North versus Southeast Asians. This observation may be due to the race differences
between North- and Southeast Asia. While plausible, such trends were not found in pseudogene regions
[Table 2]. We therefore suggest that only active genes participated in the environmental evolution or
adaptation. This is additional evidence that supports the role of geographic blocks in the evolution of HBV-
related SNPs in HLA regions.