Page 128 - Read Online
P. 128
Page 6 of 12 Tai et al. Hepatoma Res 2020;6:74 I http://dx.doi.org/10.20517/2394-5079.2020.54
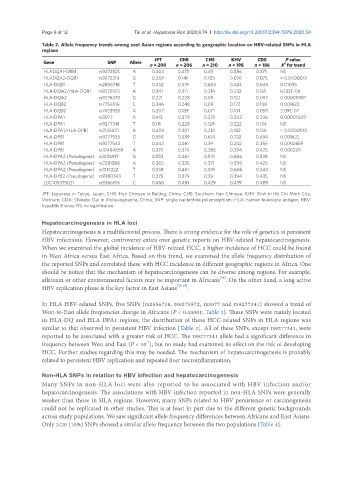
Table 2. Allele frequency trends among east Asian regions according to geographic location on HBV-related SNPs in HLA
regions
JPT CHB CHS KHV CDX P value
Gene SNP Allele 2
n = 208 n = 206 n = 210 n = 198 n = 186 X for trend
HLADQA1-DRB1 rs9272105 A 0.404 0.471 0.49 0.586 0.371 NS
HLADQA2-DQB1 rs9275319 G 0.269 0.141 0.105 0.076 0.075 < 0.0000001
HLA-DQB1 rs2856718 T 0.452 0.519 0.605 0.424 0.645 0.01035
HLA-DQA2/HLA-DQB1 rs9275572 A 0.341 0.311 0.214 0.232 0.161 6.133E-06
HLA-DQA2 rs9276370 G 0.221 0.228 0.09 0.152 0.097 0.00005987
HLA-DQB2 rs7756516 C 0.346 0.248 0.09 0.172 0.108 0.001622
HLA-DQB2 rs7453920 A 0.207 0.189 0.071 0.101 0.059 2.09E-07
HLA-DPA1 rs3077 A 0.413 0.379 0.271 0.303 0.226 0.00001509
HLA-DPA1 rs9277341 T 0.111 0.228 0.129 0.222 0.134 NS
HLA-DPA1/HLA-DPB1 rs3135021 A 0.409 0.301 0.214 0.182 0.156 < 0.0000001
HLA-DPB1 rs9277535 G 0.558 0.539 0.614 0.722 0.634 0.001622
HLA-DPB1 rs9277542 T 0.442 0.461 0.39 0.242 0.355 0.0002659
HLA-DPB1 rs10484569 A 0.375 0.374 0.386 0.394 0.425 0.000229
HLA-DPA2 (Pseudogene) rs3128917 G 0.553 0.461 0.519 0.606 0.538 NS
HLA-DPA2 (Pseudogene) rs2281388 A 0.365 0.335 0.371 0.399 0.425 NS
HLA-DPA2 (Pseudogene) rs3117222 T 0.558 0.461 0.519 0.606 0.543 NS
HLA-DPB2 (Pseudogene) rs9380343 T 0.375 0.379 0.39 0.394 0.435 NS
LOC105375021 rs9366816 C 0.466 0.481 0.429 0.439 0.489 NS
JPT: Japanese in Tokyo, Japan; CHB: Han Chinese in Beijing, China; CHS: Southern Han Chinese; KHV: Kinh in Ho Chi Minh City,
Vietnam; CDX: Chinese Dai in Xishuangbanna, China; SNP: single nucleotide polymorphism; HLA: human leukocyte antigen; HBV:
hepatitis B virus; NS: no significance
Hepatocarcinogenesis in HLA loci
Hepatocarcinogenesis is a multifactorial process. There is strong evidence for the role of genetics in persistent
HBV infections. However, controversy exists over genetic reports on HBV-related hepatocarcinogenesis.
When we examined the global incidence of HBV-related HCC, a higher incidence of HCC could be found
in West Africa versus East Africa. Based on this trend, we examined the allele frequency distribution of
the reported SNPs and correlated these with HCC incidence in different geographic regions in Africa. One
should be notice that the mechanism of hepatocarcinogenesis can be diverse among regions. For example,
[39]
aflatoxin or other environmental factors may be important in Africans . On the other hand, a long active
HBV replication phase is the key factor in East Asians [26-29] .
In HLA HBV-related SNPs, five SNPs (rs2856718, rs9275572, rs3077 and rs9277341) showed a trend of
West-to-East allele frequencies change in Africans (P < 0.00001; Table 3). These SNPs were mainly located
in HLA-DQ and HLA DPA1 regions; the distribution of these HCC-related SNPs in HLA regions was
similar to that observed in persistent HBV infection [Table 2]. All of these SNPs, except rs9277341, were
reported to be associated with a greater risk of HCC. The rs9277341 allele had a significant difference in
-7
frequency between West and East (P < 10 ), but no study had examined its effect on the risk of developing
HCC. Further studies regarding this may be needed. The mechanism of hepatocarcinogenesis is probably
related to persistent HBV replication and repeated liver necroinflammation.
Non-HLA SNPs in relation to HBV infection and hepatocarcinogenesis
Many SNPs in non-HLA loci were also reported to be associated with HBV infection and/or
hepatocarcinogenesis. The associations with HBV infection reported in non-HLA SNPs were generally
weaker than those in HLA regions. However, many SNPs related to HBV persistence or carcinogenesis
could not be replicated in other studies. This is at least in part due to the different genetic backgrounds
across study populations. We saw significant allele frequency differences between Africans and East Asians.
Only 2/20 (10%) SNPs showed a similar allele frequency between the two populations [Table 4].