Page 130 - Read Online
P. 130
Page 8 of 12 Tai et al. Hepatoma Res 2020;6:74 I http://dx.doi.org/10.20517/2394-5079.2020.54
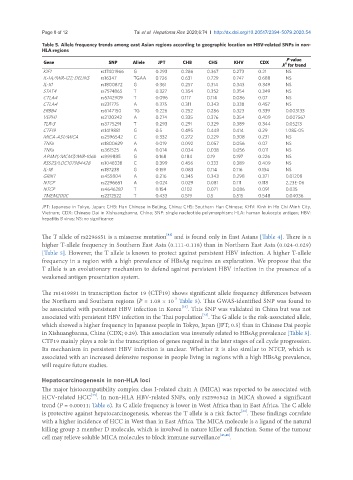
Table 5. Allele frequency trends among east Asian regions according to geographic location on HBV-related SNPs in non-
HLA regions
P value
Gene SNP Allele JPT CHB CHS KHV CDX 2
X for trend
KIF1 rs17401966 G 0.293 0.286 0.367 0.273 0.21 NS
IL-1A/MIR-122; DELINS rs16347 TGAA 0.726 0.631 0.729 0.747 0.688 NS
IL-10 rs1800872 G 0.361 0.257 0.314 0.343 0.349 NS
STAT4 rs7574865 T 0.327 0.354 0.352 0.354 0.349 NS
CTLA4 rs5742909 T 0.096 0.117 0.114 0.086 0.07 NS
CTLA4 rs231775 A 0.375 0.311 0.343 0.338 0.457 NS
ERBB4 rs6147150 TG 0.226 0.252 0.286 0.323 0.339 0.003133
VEPH1 rs2120243 A 0.274 0.335 0.376 0.354 0.409 0.007567
TLR-3 rs3775291 T 0.293 0.291 0.329 0.389 0.344 0.05213
CTF19 rs1419881 G 0.5 0.495 0.448 0.414 0.29 1.08E-05
MICA-AS1/MICA rs2596542 C 0.332 0.272 0.229 0.308 0.231 NS
TNFa rs1800629 A 0.019 0.092 0.057 0.056 0.07 NS
TNFa rs361525 A 0.014 0.034 0.038 0.056 0.011 NS
AP4M1/MCM7/MIR-106b rs999885 G 0.168 0.184 0.19 0.197 0.226 NS
RSS23/LOC107984428 rs1048338 C 0.399 0.456 0.333 0.389 0.409 NS
IL-18 rs187238 G 0.159 0.083 0.114 0.116 0.134 NS
GRIK1 rs455804 A 0.216 0.345 0.343 0.298 0.371 0.01208
NTCP rs2296651 A 0.024 0.029 0.081 0.111 0.118 2.23E-06
NTCP rs4646287 T 0.154 0.102 0.071 0.086 0.091 0.035
TMEM200C rs2212522 T 0.433 0.519 0.5 0.515 0.548 0.04036
JPT: Japanese in Tokyo, Japan; CHB: Han Chinese in Beijing, China; CHS: Southern Han Chinese; KHV: Kinh in Ho Chi Minh City,
Vietnam; CDX: Chinese Dai in Xishuangbanna, China; SNP: single nucleotide polymorphism; HLA: human leukocyte antigen; HBV:
hepatitis B virus; NS: no significance
[41]
The T allele of rs2296651 is a missense mutation and is found only in East Asians [Table 4]. There is a
higher T-allele frequency in Southern East Asia (0.111-0.118) than in Northern East Asia (0.024-0.029)
[Table 5]. However, the T allele is known to protect against persistent HBV infection. A higher T-allele
frequency in a region with a high prevalence of HBsAg requires an explanation. We propose that the
T allele is an evolutionary mechanism to defend against persistent HBV infection in the presence of a
weakened antigen presentation system.
The rs1419881 in transcription factor 19 (CTF19) shows significant allele frequency differences between
-5
the Northern and Southern regions (P = 1.08 × 10 Table 5). This GWAS-identified SNP was found to
be associated with persistent HBV infection in Korea . This SNP was validated in China but was not
[21]
[42]
associated with persistent HBV infection in the Thai population . The G allele is the risk-associated allele,
which showed a higher frequency in Japanese people in Tokyo, Japan (JPT; 0.5) than in Chinese Dai people
in Xishuangbanna, China (CDX; 0.29). This association was inversely related to HBsAg prevalence [Table 5].
CTF19 mainly plays a role in the transcription of genes required in the later stages of cell cycle progression.
Its mechanism in persistent HBV infection is unclear. Whether it is also similar to NTCP, which is
associated with an increased defensive response in people living in regions with a high HBsAg prevalence,
will require future studies.
Hepatocarcinogenesis in non-HLA loci
The major histocompatibility complex class I-related chain A (MICA) was reported to be associated with
[43]
HCV-related HCC . In non-HLA HBV-related SNPs, only rs2596542 in MICA showed a significant
trend (P = 0.00011; Table 6). Its C allele frequency is lower in West Africa than in East Africa. The C allele
[44]
is protective against hepatocarcinogenesis, whereas the T allele is a risk factor . These findings correlate
with a higher incidence of HCC in West than in East Africa. The MICA molecule is a ligand of the natural
killing group 2 member D molecule, which is involved in nature killer cell function. Some of the tumour
cell may relieve soluble MICA molecules to block immune surveillance [45,46] .