Page 28 - Read Online
P. 28
Page 630 Laubach et al. Cancer Drug Resist 2023;6:611-41 https://dx.doi.org/10.20517/cdr.2023.60
+
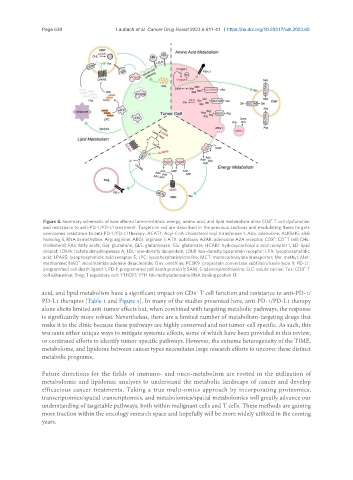
Figure 4. Summary schematic of how altered tumor-intrinsic energy, amino acid, and lipid metabolism drive CD8 T cell dysfunction
and resistance to anti-PD-1/PD-L1 treatment. Targets in red are described in the previous sections and modulating these targets
overcomes resistance to anti-PD-1/PD-L1 therapy. ACAT1: Acyl-CoA cholesterol acyl transferase 1; Ado: adenosine; ALKBH5: alkB
+
+
homolog 5, RNA demethylase; Arg: arginine; ARG1: arginase 1; ATX: autotaxin; A2AR: adenosine A2A receptor; CD8 : CD T cell; CHL:
cholesterol; FAs: fatty acids; Gln: glutamine; GLS: glutaminase; Glu: glutamate; HCAR1: hydroxycarboxylic acid receptor 1; LD: lipid
droplet; LDHA: lactate dehydrogenase A; LDL: low-density lipoprotein; LDLR: low-density lipoprotein receptor; LPA: lysophosphatidic
acid; LPAR5: lysophosphatidic acid receptor 5; LPC: lysophosphatidylcholine; MCT: monocarboxylate transporter; Me: methyl; Met:
+
methionine; NAD : nicotinamide adenine dinucleotide; Orn: ornithine; PCSK9: proprotein convertase subtilisin/kexin type 9; PD-L1:
+
programmed cell death ligand 1; PD-1: programmed cell death protein 1; SAM: S-adenosylmethionine; SLC: solute carrier; Tex: CD8 T
cell exhaustion; Treg: T regulatory cell; YTHDF1: YTH N6-methyladenosine RNA binding protein F1.
acid, and lipid metabolism have a significant impact on CD8 T cell function and resistance to anti-PD-1/
+
PD-L1 therapies [Table 1 and Figure 4]. In many of the studies presented here, anti-PD-1/PD-L1 therapy
alone elicits limited anti-tumor effects but, when combined with targeting metabolic pathways, the response
is significantly more robust. Nevertheless, there are a limited number of metabolism-targeting drugs that
make it to the clinic because these pathways are highly conserved and not tumor-cell specific. As such, this
warrants either unique ways to mitigate systemic effects, some of which have been provided in this review,
or continued efforts to identify tumor-specific pathways. However, the extreme heterogeneity of the TIME,
metabolome, and lipidome between cancer types necessitates large research efforts to uncover these distinct
metabolic programs.
Future directions for the fields of immuno- and onco-metabolism are rooted in the utilization of
metabolomic and lipidomic analyses to understand the metabolic landscape of cancer and develop
efficacious cancer treatments. Taking a true multi-omics approach by incorporating proteomics,
transcriptomics/spatial transcriptomics, and metabolomics/spatial metabolomics will greatly advance our
understanding of targetable pathways, both within malignant cells and T cells. These methods are gaining
more traction within the oncology research space and hopefully will be more widely utilized in the coming
years.