Page 14 - Read Online
P. 14
Dong et al. UPI peptide as cancer therapeutic
A B
C D
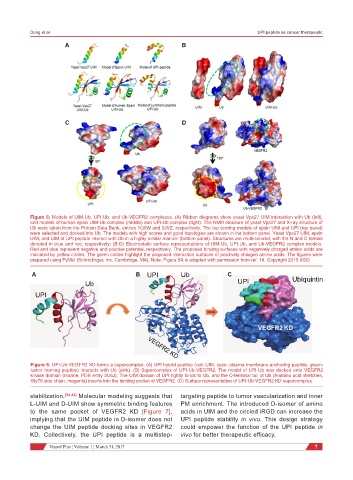
Figure 5: Models of UIM-Ub, UPI-Ub, and Ub-VEGFR2 complexes. (A) Ribbon diagrams show yeast Vps27 UIM interaction with Ub (left),
and models of human epsin UIM-Ub complex (middle) and UPI-Ub complex (right). The NMR structure of yeast Vps27 and X-ray structure of
Ub were taken from the Protein Data Bank, entries 1Q0W and 3JVZ, respectively. The top scoring models of epsin UIM and UPI (top panel)
were selected and docked into Ub. The models with high scores and good topologies are shown in the bottom panel. Yeast Vps27 UIM, epsin
UIM, and UIM of UPI peptide interact with Ub in a highly similar manner (bottom panel). Structures are multi-colored, with the N and C termini
denoted in blue and red, respectively; (B-D) Electrostatic surface representations of UIM-Ub, UPI-Ub, and Ub-VEGFR2 complex models.
Red and blue represent negative and positive potential, respectively. The proposed binding surfaces with negatively charged amino acids are
indicated by yellow circles. The green circles highlight the proposed interaction surfaces of positively charged amino acids. The figures were
prepared using PyMol (Schrödinger, Inc, Cambridge, MA). Note: Figure 5A is adapted with permission from ref. 19. Copyright 2015 ASCI
A B UPI Ub C Ubiquintin
Ub UPI
UPI
VEGFR2 KD
Figure 6: UPI-Ub-VEGFR2 KD forms a supercomplex. (A) UPI hybrid peptide (red: UIM, cyan: plasma membrane-anchoring peptide, green:
tumor homing peptide) interacts with Ub (pink). (B) Supercomplex of UPI-Ub-VEGFR2. The model of UPI-Ub was docked onto VEGFR2
kinase domain (marine, PDB entry 3U6J). The UIM domain of UPI tightly binds to Ub, and the C-terminal tail of Ub (8-amino acid stretches,
Gly76 side chain, magenta) inserts into the binding pocket of VEGFR2. (C) Surface representation of UPI-Ub-VEGFR2 KD supercomplex
stabilization. [30-32] Molecular modeling suggests that targeting peptide to tumor vascularization and inner
L-UIM and D-UIM show symmetric binding features PM enrichment. The introduced D-isomer of amino
to the same pocket of VEGFR2 KD [Figure 7], acids in UIM and the circled iRGD can increase the
implying that the UIM peptide in D-isomer does not UPI peptide stability in vivo. This design strategy
change the UIM peptide docking sites in VEGFR2 could empower the function of the UPI peptide in
KD. Collectively, the UPI peptide is a multistep- vivo for better therapeutic efficacy.
Vessel Plus ¦ Volume 1 ¦ March 31, 2017 7