Page 65 - Read Online
P. 65
Feusier et al. J Transl Genet Genom 2021;5:189-99 https://dx.doi.org/10.20517/jtgg.2021.05 Page 195
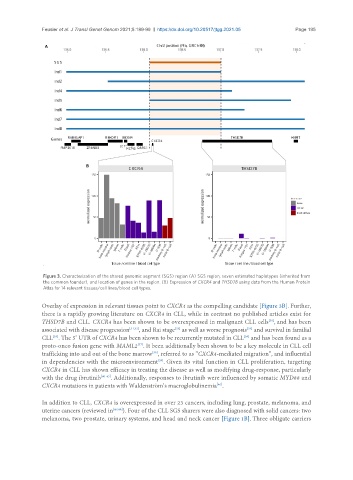
Figure 3. Characterization of the shared genomic segment (SGS) region (A) SGS region, seven estimated haplotypes (inherited from
the common founder), and location of genes in the region. (B) Expression of CXCR4 and THSD7B using data from the Human Protein
Atlas for 14 relevant tissues/cell lines/blood cell types.
Overlay of expression in relevant tissues point to CXCR4 as the compelling candidate [Figure 3B]. Further,
there is a rapidly growing literature on CXCR4 in CLL, while in contrast no published articles exist for
[30]
THSD7B and CLL. CXCR4 has been shown to be overexpressed in malignant CLL cells , and has been
[33]
associated with disease progression [31,32] , and Rai stage as well as worse prognosis and survival in familial
[34]
[36]
CLL . The 5’ UTR of CXCR4 has been shown to be recurrently mutated in CLL and has been found as a
[35]
proto-onco fusion gene with MAML2 . It been additionally been shown to be a key molecule in CLL cell
[37]
trafficking into and out of the bone marrow , referred to as “CXCR4-mediated migration”, and influential
[38]
in dependencies with the microenvironment . Given its vital function in CLL proliferation, targeting
[39]
CXCR4 in CLL has shown efficacy in treating the disease as well as modifying drug-response, particularly
with the drug ibrutinib [40-45] . Additionally, responses to ibrutinib were influenced by somatic MYD88 and
[46]
CXCR4 mutations in patients with Waldenström’s macroglobulinemia .
In addition to CLL, CXCR4 is overexpressed in over 23 cancers, including lung, prostate, melanoma, and
uterine cancers (reviewed in [47,48] ). Four of the CLL SGS sharers were also diagnosed with solid cancers: two
melanoma, two prostate, urinary systems, and head and neck cancer [Figure 1B]. Three obligate carriers