Page 64 - Read Online
P. 64
Page 194 Feusier et al. J Transl Genet Genom 2021;5:189-99 https://dx.doi.org/10.20517/jtgg.2021.05
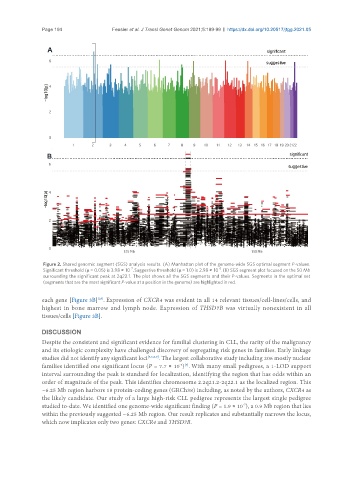
Figure 2. Shared genomic segment (SGS) analysis results. (A) Manhattan plot of the genome-wide SGS optimal segment P-values.
-6
-7
Significant threshold (μ = 0.05) is 3.98 × 10 . Suggestive threshold (μ = 1.0) is 2.98 × 10 . (B) SGS segment plot focused on the 50 Mb
surrounding the significant peak at 2q22.1. The plot shows all the SGS segments and their P-values. Segments in the optimal set
(segments that are the most significant P-value at a position in the genome) are highlighted in red.
each gene [Figure 3B] . Expression of CXCR4 was evident in all 14 relevant tissues/cell-lines/cells, and
[28]
highest in bone marrow and lymph node. Expression of THSD7B was virtually nonexistent in all
tissues/cells [Figure 3B].
DISCUSSION
Despite the consistent and significant evidence for familial clustering in CLL, the rarity of the malignancy
and its etiologic complexity have challenged discovery of segregating risk genes in families. Early linkage
studies did not identify any significant loci [5,6,8,9] . The largest collaborative study including 206 mostly nuclear
families identified one significant locus (P = 7.7 × 10 ) . With many small pedigrees, a 1-LOD support
-5 [7]
interval surrounding the peak is standard for localization, identifying the region that has odds within an
order of magnitude of the peak. This identifies chromosome 2.2q21.2-2q22.1 as the localized region. This
~6.25 Mb region harbors 18 protein-coding genes (GRCh38) including, as noted by the authors, CXCR4 as
the likely candidate. Our study of a large high-risk CLL pedigree represents the largest single pedigree
-7
studied to-date. We identified one genome-wide significant finding (P = 1.9 × 10 ), a 0.9 Mb region that lies
within the previously suggested ~6.25 Mb region. Our result replicates and substantially narrows the locus,
which now implicates only two genes: CXCR4 and THSD7B.