Page 19 - Read Online
P. 19
Page 8 Donskov et al. J Transl Genet Genom 2021;5:136-62 https://dx.doi.org/10.20517/jtgg.2021.12
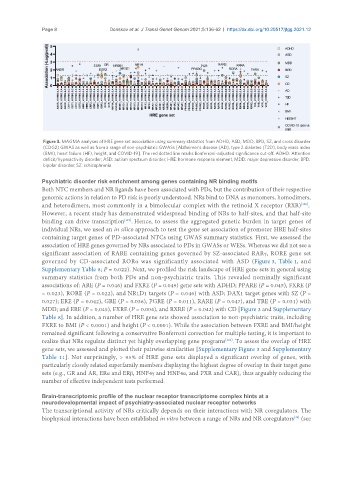
Figure 3. MAGMA analyses of HRE gene set association using summary statistics from ADHD, ASD, MDD, BPD, SZ, and cross disorder
(CDG2) GWAS as well as from a range of non-psychiatric GWASs [Alzheimer’s disease (AD), type 2 diabetes (T2D), body mass index
(BMI), heart failure (HF), height, and COVID-19]. The red dotted line marks Bonferroni-adjusted significance cut-off. ADHD: Attention
deficit/hyperactivity disorder; ASD: autism spectrum disorder; HRE: hormone response element; MDD: major depressive disorder; BPD:
bipolar disorder; SZ: schizophrenia.
Psychiatric disorder risk enrichment among genes containing NR binding motifs
Both NTC members and NR ligands have been associated with PDs, but the contribution of their respective
genomic actions in relation to PD risk is poorly understood. NRs bind to DNA as monomers, homodimers,
and heterodimers, most commonly in a bimolecular complex with the retinoid X receptor (RXR) .
[120]
However, a recent study has demonstrated widespread binding of NRs to half-sites, and that half-site
binding can drive transcription . Hence, to assess the aggregated genetic burden in target genes of
[121]
individual NRs, we used an in silico approach to test the gene set association of promotor HRE half-sites
containing target genes of PD-associated NTCs using GWAS summary statistics. First, we assessed the
association of HRE genes governed by NRs associated to PDs in GWASs or WESs. Whereas we did not see a
significant association of RARE containing genes governed by SZ-associated RARγ, RORE gene set
governed by CD-associated RORα was significantly associated with ASD (Figure 3, Table 1, and
Supplementary Table 8; P = 0.022). Next, we profiled the risk landscape of HRE gene sets in general using
summary statistics from both PDs and non-psychiatric traits. This revealed nominally significant
associations of: ARE (P = 0.046) and FXRE (P = 0.049) gene sets with ADHD; PPARE (P = 0.045), FXRE (P
= 0.023), RORE (P = 0.022), and NR1D1 targets (P = 0.046) with ASD; DAX1 target genes with SZ (P =
0.027); ERE (P = 0.042), GRE (P = 0.036), PGRE (P = 0.011), RARE (P = 0.047), and TRE (P = 0.031) with
MDD; and ERE (P = 0.045), FXRE (P = 0.004), and RXRE (P = 0.042) with CD [Figure 3 and Supplementary
Table 8]. In addition, a number of HRE gene sets showed association to non-psychiatric traits, including
FXRE to BMI (P < 0.0001) and height (P < 0.0001). While the association between FXRE and BMI/height
remained significant following a conservative Bonferroni correction for multiple testing, it is important to
realize that NRs regulate distinct yet highly overlapping gene programs . To assess the overlap of HRE
[121]
gene sets, we assessed and plotted their pairwise similarities [Supplementary Figure 3 and Supplementary
Table 11]. Not surprisingly, > 95% of HRE gene sets displayed a significant overlap of genes, with
particularly closely related superfamily members displaying the highest degree of overlap in their target gene
sets (e.g., GR and AR, ERα and ERβ, HNF4γ and HNF4α, and PXR and CAR), thus arguably reducing the
number of effective independent tests performed.
Brain-transcriptomic profile of the nuclear receptor transcriptome complex hints at a
neurodevelopmental impact of psychiatry-associated nuclear receptor networks
The transcriptional activity of NRs critically depends on their interactions with NR coregulators. The
biophysical interactions have been established in vitro between a range of NRs and NR coregulators (see
[49]