Page 18 - Read Online
P. 18
Donskov et al. J Transl Genet Genom 2021;5:136-62 https://dx.doi.org/10.20517/jtgg.2021.12 Page 7
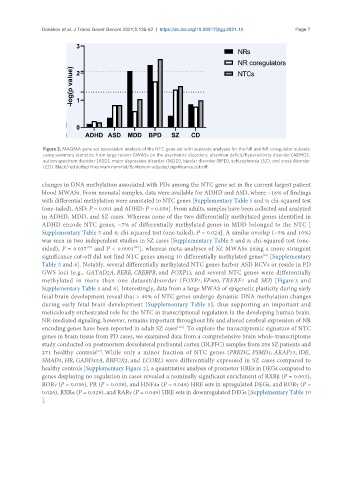
Figure 2. MAGMA gene set association analysis of the NTC gene set with separate analyses for the NR and NR coregulator subsets
using summary statistics from large recent GWASs on the psychiatric disorders: attention deficit/hyperactivity disorder (ADHD),
autism spectrum disorder (ASD), major depressive disorder (MDD), bipolar disorder (BPD), schizophrenia (SZ), and cross disorder
(CD). Black/red dotted lines mark nominal/Bonferroni-adjusted significance cut-off.
changes in DNA methylation associated with PDs among the NTC gene set in the current largest patient
blood MWASs. From neonatal samples, data were available for ADHD and ASD, where ~16% of findings
with differential methylation were annotated to NTC genes [Supplementary Table 5 and 9; chi-squared test
(one-tailed), ASD: P = 0.001 and ADHD: P = 0.050]. From adults, samples have been collected and analyzed
in ADHD, MDD, and SZ cases. Whereas none of the two differentially methylated genes identified in
ADHD encode NTC genes, ~7% of differentially methylated genes in MDD belonged to the NTC [
Supplementary Table 5 and 8; chi-squared test (one-tailed), P = 0.024]. A similar overlap (~5% and 10%)
was seen in two independent studies in SZ cases [Supplementary Table 5 and 8; chi-squared test (one-
[79]
[80]
sided), P = 0.037 and P < 0.0001 ], whereas meta-analyses of SZ MWASs using a more stringent
significance cut-off did not find NTC genes among 10 differentially methylated genes [Supplementary
[81]
Table 5 and 8]. Notably, several differentially methylated NTC genes harbor ASD RCVs or reside in PD
GWS loci (e.g., GATAD2A, RERE, CREBPB, and FOXP1), and several NTC genes were differentially
methylated in more than one dataset/disorder (FOXP1, EP400, TRERF1 and SKI) [Figure 3 and
Supplementary Table 5 and 8]. Interestingly, data from a large MWAS of epigenetic plasticity during early
fetal brain development reveal that > 40% of NTC genes undergo dynamic DNA methylation changes
during early fetal brain development [Supplementary Table 5], thus supporting an important and
meticulously orchestrated role for the NTC in transcriptional regulation in the developing human brain.
NR-mediated signaling, however, remains important throughout life and altered cerebral expression of NR
encoding genes have been reported in adult SZ cases . To explore the transcriptomic signature of NTC
[119]
genes in brain tissue from PD cases, we examined data from a comprehensive brain whole-transcriptome
study conducted on postmortem dorsolateral prefrontal cortex (DLPFC) samples from 258 SZ patients and
271 healthy controls . While only a minor fraction of NTC genes (PRKDC, PSMD1, AKAP13, IDE,
[92]
SMAD3, HR, GADD45A, RBFOX2, and LCORL) were differentially expressed in SZ cases compared to
healthy controls [Supplementary Figure 2], a quantitative analysis of promotor HREs in DEGs compared to
genes displaying no regulation in cases revealed a nominally significant enrichment of RXRβ (P = 0.003),
RORγ (P = 0.036), PR (P = 0.038), and HNF4α (P = 0.048) HRE sets in upregulated DEGs, and RORγ (P =
0.026), RXRα (P = 0.028), and RARγ (P = 0.049) HRE sets in downregulated DEGs [Supplementary Table 10
].