Page 15 - Read Online
P. 15
Page 4 Donskov et al. J Transl Genet Genom 2021;5:136-62 https://dx.doi.org/10.20517/jtgg.2021.12
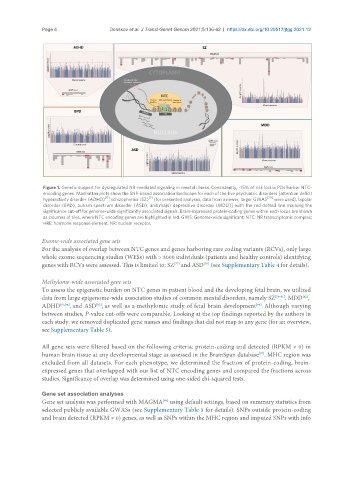
Figure 1. Genetic support for dysregulated NR-mediated signaling in mental illness. Consistently, ~15% of risk loci in PDs harbor NTC-
encoding genes. Manhattan plots show the SNP-based association landscape for each of the five psychiatric disorders [attention deficit
[11]
[7]
hyperactivity disorder (ADHD) , schizophrenia (SZ) (for presented analyses, data from a newer, larger GWAS [70] were used), bipolar
disorder (BPD), autism spectrum disorder (ASD), and major depressive disorder (MDD)] with the red dotted line marking the
significance cut-off for genome-wide significantly associated signals. Brain-expressed protein-coding genes within each locus are shown
as columns of tiles, where NTC encoding genes are highlighted in red. GWS: Genome-wide significant; NTC: NR transcriptomic complex;
HRE: hormone response element; NR: nuclear receptor.
Exome-wide associated gene sets
For the analysis of overlap between NTC genes and genes harboring rare coding variants (RCVs), only large
whole exome sequencing studies (WESs) with > 3000 individuals (patients and healthy controls) identifying
[77]
[78]
genes with RCVs were assessed. This is limited to: SZ and ASD (see Supplementary Table 4 for details).
Methylome-wide associated gene sets
To assess the epigenetic burden on NTC genes in patient blood and the developing fetal brain, we utilized
data from large epigenome-wide association studies of common mental disorders, namely SZ [79-81] , MDD ,
[82]
ADHD [83,84] , and ASD , as well as a methylomic study of fetal brain development . Although varying
[85]
[86]
between studies, P-value cut-offs were comparable. Looking at the top findings reported by the authors in
each study, we removed duplicated gene names and findings that did not map to any gene (for an overview,
see Supplementary Table 5).
All gene sets were filtered based on the following criteria: protein-coding and detected (RPKM ≠ 0) in
human brain tissue at any developmental stage as assessed in the BrainSpan database . MHC region was
[87]
excluded from all datasets. For each phenotype, we determined the fraction of protein-coding, brain-
expressed genes that overlapped with our list of NTC encoding genes and compared the fractions across
studies. Significance of overlap was determined using one-sided chi-squared tests.
Gene set association analyses
Gene set analysis was performed with MAGMA using default settings, based on summary statistics from
[88]
selected publicly available GWASs (see Supplementary Table 3 for details). SNPs outside protein-coding
and brain detected (RPKM ≠ 0) genes, as well as SNPs within the MHC region and imputed SNPs with info