Page 192 - Read Online
P. 192
Glinsky Genetic signatures of lethal disease in early stage prostate cancer
lethal prostate cancer at diagnosis. We applied the identify lethal disease in prostate cancer patients of
univariate Cox regression analysis to the entire cohort differing ages. Remarkably, Kaplan-Meier survival
of 281 patients to identify several GES with the P value analysis has determined that 98 genes GES
< 0.01 which appear to perform better than the best
clinico-pathological co-variate, Gleason score (P = performed very efficiently in stratification of prostate
0.0113; Supplemental Table 1). Most of these GES
outperformed the clinico-pathological classification
model in multivariate Cox regression analysis as well
[Supplemental Table 2].
Separating the cohort of 281 patients into training
and test cohorts and using the Kaplan-Meier survival
analysis, we identified 98 genes GES that manifest
the highly significant classification performance in the
training set, retained highly consistent classification
performance in the test set, and remained a highly
significant classifier in the pooled cohort [Figure 1].
It is important to note that in all secondary validation
screens following the training set analysis no further
adjustments to the threshold selection and classification
protocols were made.
Notably, prostate cancer patients with identical
Gleason scores (e.g. Gleason 6 patients and Gleason
7 patients) which were segregated into lethal and
moderate disease sub-groups based on 98 genes
GES classification had highly significant differences
in the survival rates [Figure 1]. These data suggest
that 98 genes GES may be useful in identifying lethal
disease in patients diagnosed with low grade localized
prostate cancer [Supplemental Table 3]. To test this
hypothesis, we performed Kaplan-Meier survival
analysis based on 98 genes GES classification
in the cohort of 200 patients with Gleason 6 and 7
prostate cancer [Figure 2]. We found that 98 genes
GES is a highly significant classifier of Gleason 6
and 7 prostate cancer patients into sub-groups with
lethal and moderate disease [Figure 2]. Ninety-eight
genes GES of lethal prostate cancer performs as a
highly significant after segregation of patients into
separate Gleason 6 and Gleason 7 sub-groups: 89%
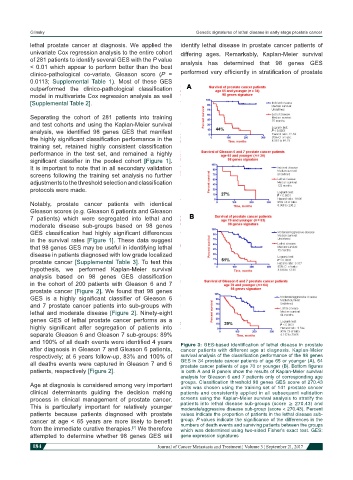
and 100% of all death events were identified 4 years Figure 3: GES-based identification of lethal disease in prostate
after diagnosis in Gleason 7 and Gleason 6 patients, cancer patients with different age at diagnosis. Kaplan-Meier
respectively; at 6 years follow-up, 83% and 100% of survival analysis of the classification performance of the 98 genes
all deaths events were captured in Gleason 7 and 6 GES in 34 prostate cancer patients of age 65 or younger (A), 64
prostate cancer patients of age 70 or younger (B). Bottom figures
patients, respectively [Figure 2]. in both A and B panels show the results of Kaplan-Meier survival
analysis for Gleason 6 and 7 patients only of corresponding age
Age at diagnosis is considered among very important groups. Classification threshold 98 genes GES score of 270.43
units was chosen using the training set of 141 prostate cancer
clinical determinants guiding the decision making patients and consistently applied in all subsequent validation
process in clinical management of prostate cancer. screens using the Kaplan-Meier survival analysis to stratify the
This is particularly important for relatively younger patients into lethal disease sub-groups (score ≥ 270.43) and
moderate/aggressive disease sub-group (score < 270.43). Percent
patients because patients diagnosed with prostate values indicate the proportion of patients in the lethal disease sub-
cancer at age < 65 years are more likely to benefit group. P values indicate the significance of the differences in the
numbers of death events and surviving patients between the groups
from the immediate curative therapies. We therefore which was determined using two-sided Fisher’s exact test. GES:
[7]
attempted to determine whether 98 genes GES will gene expression signatures
184 Journal of Cancer Metastasis and Treatment ¦ Volume 3 ¦ September 21, 2017