Page 282 - Read Online
P. 282
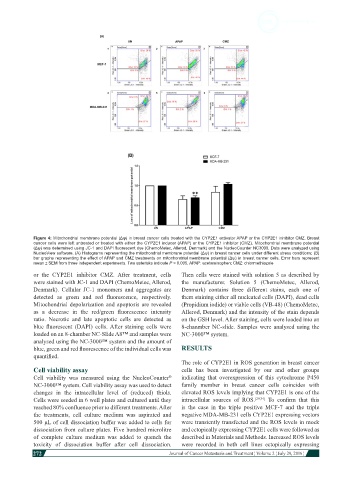
Figure 4: Mitochondrial membrane potential (Δѱ) in breast cancer cells treated with the CYP2E1 activator APAP or the CYP2E1 inhibitor CMZ. Breast
cancer cells were left untreated or treated with either the CYP2E1 inducer (APAP) or the CYP2E1 inhibitor (CMZ). Mitochondrial membrane potential
(Δѱ) was determined using JC-1 and DAPI fluorescent dye (ChemoMetec, Allerod, Denmark) and the NucleoCounter NC3000. Data were analyzed using
NucleoView software. (A) Histograms representing the mitochondrial membrane potential (Δѱ) in breast cancer cells under different stress conditions; (B)
bar graphs representing the effect of APAP and CMZ treatments on mitochondrial membrane potential (Δѱ) in breast cancer cells. Error bars represent
mean ± SEM from three independent experiments. Two asterisks indicate P < 0.005. APAP: acetaminophen; CMZ: chlormethiazole
or the CYP2E1 inhibitor CMZ. After treatment, cells Then cells were stained with solution 5 as described by
were stained with JC-1 and DAPI (ChemoMetec, Allerod, the manufacturer. Solution 5 (ChemoMetec, Allerod,
Denmark). Cellular JC-1 monomers and aggregates are Denmark) contains three different stains, each one of
detected as green and red fluorescence, respectively. them staining either all nucleated cells (DAPI), dead cells
Mitochondrial depolarization and apoptosis are revealed (Propidium iodide) or viable cells (VB-48) (ChemoMetec,
as a decrease in the red/green fluorescence intensity Allerod, Denmark) and the intensity of the stain depends
ratio. Necrotic and late apoptotic cells are detected as on the GSH level. After staining, cells were loaded into an
blue fluorescent (DAPI) cells. After staining cells were 8-chanmber NC-slide. Samples were analysed using the
loaded on an 8-chamber NC-Slide A8™ and samples were NC-3000™ system.
analysed using the NC-3000™ system and the amount of
blue, green and red fluorescence of the individual cells was RESULTS
quantified.
The role of CYP2E1 in ROS generation in breast cancer
Cell viability assay cells has been investigated by our and other groups
Cell viability was measured using the NucleoCounter indicating that overexpression of this cytochrome P450
®
NC-3000™ system. Cell viability assay was used to detect family member in breast cancer cells coincides with
changes in the intracellular level of (reduced) thiols. elevated ROS levels implying that CYP2E1 is one of the
Cells were seeded in 6 well plates and cultured until they intracellular sources of ROS. [29,34] To confirm that this
reached 80% confluence prior to different treatments. After is the case in the triple positive MCF-7 and the triple
the treatments, cell culture medium was aspirated and negative MDA-MB-231 cells CYP2E1 expressing vectors
500 μL of cell dissociation buffer was added to cells for were transiently transfected and the ROS levels in mock
dissociation from culture plates. Five hundred microlitre and ectopically expressing CYP2E1 cells were followed as
of complete culture medium was added to quench the described in Materials and Methods. Increased ROS levels
toxicity of dissociation buffer after cell dissociation. were recorded in both cell lines ectopically expressing
272
Journal of Cancer Metastasis and Treatment ¦ Volume 2 ¦ July 29, 2016 ¦