Page 59 - Read Online
P. 59
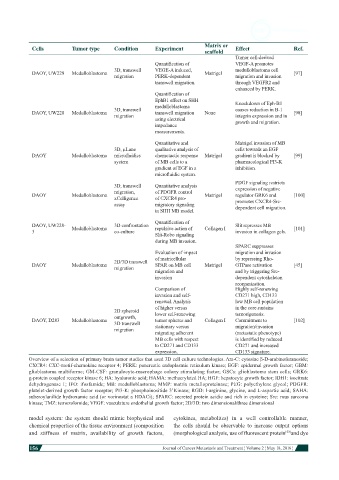
Matrix or
Cells Tumor type Condition Experiment scaffold Effect Ref.
Tumor cell-derived
Quantification of VEGF-A promotes
3D, transwell VEGE-A induced, medulloblastoma cell
DAOY, UW228 Medulloblastoma Matrigel [97]
migration PERK-dependent migration and invasion
transwell migration. through VEGFR2 and
enhanced by PERK.
Quantification of
EphB1 effect on SHH Knockdown of Eph-B1
medulloblastoma
3D, transwell causes reduction in B-1
DAOY, UW228 Medulloblastoma transwell migration None [98]
migration integrin expression and in
using electrical growth and migration.
impedance
measurements.
Quantitative and Matrigel invasion of MB
3D, µLane qualitative analysis of cells towards an EGF
DAOY Medulloblastoma microfluidics chemotactic response Matrigel gradient is blocked by [99]
system of MB cells to a pharmacological PI3-K
gradient of EGF in a inhibition.
microfluidic system.
PDGF signaling restricts
3D, transwell Quantitative analysis expression of negative
migration, of PDGFR control
DAOY Medulloblastoma Matrigel regulator GRK6 and [100]
xCelligence of CXCR4 pro- promotes CXCR4-Src-
assay migratory signaling dependent cell migration.
in SHH MB model.
Quantification of
DAOY, UW228- 3D confrontation Slit represses MB
3 Medulloblastoma co-culture repulsive action of Collagen I invasion in collagen gels. [101]
Slit-Robo signaling
during MB invasion.
SPARC suppresses
Evaluation of impact migration and invasion
of matricellular by repressing Rho-
2D/3D transwell
DAOY Medulloblastoma SPAR on MB cell Matrigel GTPase activation [45]
migration
migration and and by triggering Src-
invasion dependent cytoskeleton
reorganization.
Comparison of Highly self-renewing
invasion and self- CD271 high, CD133
renewal. Analysis low MB cell population
of higher versus in the core sustains
2D spheroid
outgrowth, lower self-renewing tumorigenesis.
DAOY, D283 Medulloblastoma tumor spheres and Collagen I Commitment to [102]
3D transwell
migration stationary versus migration/invasion
migrating adherent (metastatic phenotype)
MB cells with respect is identified by reduced
to CD271 and CD133 CD271 and increased
expression. CD133 signature.
Overview of a selection of primary brain tumor studies that used 3D cell culture technologies. Ara-C: cytosine β-D-arabinofuranoside;
CXCR4: CXC-motif-chemokine receptor 4; PERK: pancreatic endoplasmic reticulum kinase; EGF: epidermal growth factor; GBM:
glioblastoma multiforme; GM-CSF: granulocyte-macrophage colony stimulating factor; GSCs: glioblastoma stem cells; GRK6:
g-protein coupled receptor kinase 6; HA: hyaluronic acid; HAMA: methacrylated HA; HGF: hepatocyte growth factor; IDH1: isocitrate
dehydrogenase 1; IFO: ifosfamide; MB: medulloblastoma; MMP: matrix metalloproteinase; PEG: polyethylene glycol; PDGFR:
platelet-derived growth factor receptor; PI3-K: phosphoinositide 3’Kinase; RGD: l-arginine, glycine, and L-aspartic acid; SAHA:
suberoylanilide hydroxamic acid (or vorinostat a HDACi); SPARC: secreted protein acidic and rich in cysteine; Src: rous sarcoma
kinase; TMZ: temozolomide; VEGF: vasculature endothelial growth factor; 2D/3D: two dimensional/three dimensional
model system: the system should mimic biophysical and cytokines, metabolites) in a well controllable manner,
chemical properties of the tissue environment (composition the cells should be observable to increase output options
and stiffness of matrix, availability of growth factors, (morphological analysis, use of fluorescent protein and dye
[53]
156
Journal of Cancer Metastasis and Treatment ¦ Volume 2 ¦ May 18, 2016 ¦