Page 72 - Read Online
P. 72
Pippione et al. Steroidogenic enzymes in prostate cancer
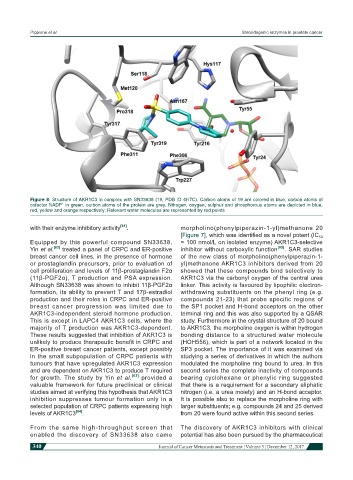
Figure 8: Structure of AKR1C3 in complex with SN33638 (19, PDB ID 4H7C). Carbon atoms of 19 are colored in blue, carbon atoms of
+
cofactor NADP in green, carbon atoms of the protein are grey. Nitrogen, oxygen, sulphur and phosphorous atoms are depicted in blue,
red, yellow and orange respectively. Relevant water molecules are represented by red points
with their enzyme inhibitory activity [61] . morpholino(phenylpiperazin-1-yl)methanone 20
[Figure 7], which was identified as a novel potent (IC 50
Equipped by this powerful compound SN33638, = 100 nmol/L on isolated enzyme) AKR1C3-selective
Yin et al. [63] treated a panel of CRPC and ER-positive inhibitor without carboxylic function [65] . SAR studies
breast cancer cell lines, in the presence of hormone of the new class of morpholino(phenylpiperazin-1-
or prostaglandin precursors, prior to evaluation of yl)methanone AKR1C3 inhibitors derived from 20
cell proliferation and levels of 11β-prostaglandin F2α showed that these compounds bind selectively to
(11β-PGF2α), T production and PSA expression. AKR1C3 via the carbonyl oxygen of the central urea
Although SN33638 was shown to inhibit 11β-PGF2α linker. This activity is favoured by lipophilic electron-
formation, its ability to prevent T and 17β-estradiol withdrawing substituents on the phenyl ring (e.g.
production and their roles in CRPC and ER-positive compounds 21-23) that probe specific regions of
breast cancer progression was limited due to the SP1 pocket and H-bond acceptors on the other
AKR1C3-independent steroid hormone production. terminal ring and this was also supported by a QSAR
This is except in LAPC4 AKR1C3 cells, where the study. Furthermore in the crystal structure of 20 bound
majority of T production was AKR1C3-dependent. to AKR1C3, the morpholine oxygen is within hydrogen
These results suggested that inhibition of AKR1C3 is bonding distance to a structured water molecule
unlikely to produce therapeutic benefit in CRPC and (HOH556), which is part of a network located in the
ER-positive breast cancer patients, except possibly SP3 pocket. The importance of it was examined via
in the small subpopulation of CRPC patients with studying a series of derivatives in which the authors
tumours that have upregulated AKR1C3 expression modulated the morpholine ring bound to urea. In this
and are dependent on AKR1C3 to produce T required second series the complete inactivity of compounds
for growth. The study by Yin et al. [63] provided a bearing cyclohexane or phenylic ring suggested
valuable framework for future preclinical or clinical that there is a requirement for a secondary aliphatic
studies aimed at verifying this hypothesis that AKR1C3 nitrogen (i.e. a urea moiety) and an H-bond acceptor.
inhibition suppresses tumour formation only in a It is possible also to replace the morpholine ring with
selected population of CRPC patients expressing high larger substituents; e.g. compounds 24 and 25 derived
levels of AKR1C3 [64] . from 20 were found active within this second series.
From the same high-throughput screen that The discovery of AKR1C3 inhibitors with clinical
enabled the discovery of SN33638 also came potential has also been pursued by the pharmaceutical
340 Journal of Cancer Metastasis and Treatment ¦ Volume 3 ¦ December 12, 2017