Page 49 - Read Online
P. 49
Warawdekar et al. CTCs from patients with metastatic breast cancer
and Table 2 show the number of CTCs identified in
patient samples and normal controls. Double-positive
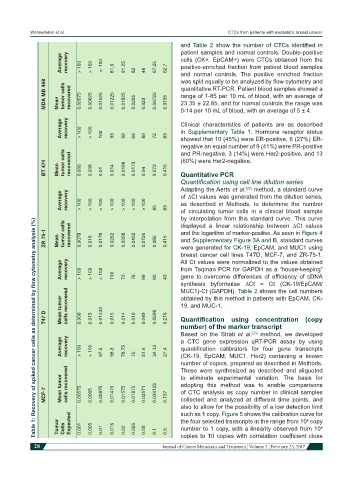
Average recovery > 100 > 100 ≈ 100 81.6 91.25 82 46 67.25 62.7 cells (CK+, EpCAM+) were CTCs obtained from the
positive-enriched fraction from patient blood samples
and normal controls. The positive enriched fraction
MDA MB 468 Mean tumor cells recovered 0.00575 0.00825 0.01025 0.01225 0.01825 0.0205 0.023 0.06725 0.3135 was split equally to be analyzed by flow cytometry and
quantitative RT-PCR. Patient blood samples showed a
range of 1-85 per 10 mL of blood, with an average of
23.35 ± 22.85, and for normal controls the range was
0-14 per 10 mL of blood, with an average of 5 ± 4.
Average recovery > 100 > 100 100 93 99 69 80 72 83 Clinical characteristics of patients are as described
in Supplementary Table 1. Hormone receptor status
showed that 10 (45%) were ER-positive, 6 (27%) ER-
negative an equal number of 9 (41%) were PR-positive
and PR-negative, 3 (14%) were Her2-positive, and 13
BT 474 Mean tumor cells recovered 0.006 0.008 0.01 0.014 0.0198 0.0173 0.04 0.072 0.415 (60%) were Her2-negative.
Quantitative PCR
Quantification using cell line dilution series
Adapting the Aerts et al. method, a standard curve
[22]
Average recovery > 100 > 100 > 100 > 100 > 100 > 100 > 100 85 83 of ∆Ct values was generated from the dilution series,
as described in Methods, to determine the number
of circulating tumor cells in a clinical blood sample
by interpolation from this standard curve. This curve
Table 1: Recovery of spiked cancer cells as determined by flow cytometry analysis (%)
displayed a linear relationship between ∆Ct values
ZR 75-1 Mean tumor cells recovered 0.0078 0.016 0.0178 0.0262 0.0328 0.0402 0.0726 0.085 0.415 and the logarithm of marker-positive. As seen in Figure 4
and Supplementary Figure 3A and B, standard curves
were generated for CK-19, EpCAM, and MUC1 using
breast cancer cell lines T47D, MCF-7, and ZR-75-1.
All Ct values were normalized to the values obtained
Average recovery > 100 > 100 > 100 100 from Taqman PCR for GAPDH as a “house-keeping”
gene to overcome differences of efficiency of cDNA
60
99
76
43
70
synthesis byformulae ∆Ct = Ct (CK-19/EpCAM/
MUC1)-Ct (GAPDH). Table 2 shows the cell numbers
Mean tumor cells recovered obtained by this method in patients with EpCAM, CK-
19, and MUC-1.
T47 D 0.006 0.015 0.01433 0.015 0.014 0.019 0.049 0.0596 0.216 Quantification using concentration (copy
number) of the marker transcript
Based on the Strati et al. method, we developed
[23]
Average recovery > 100 > 100 97.5 98.3 78.75 75 51.4 34.13 27.4 a CTC gene expression qRT-PCR assay by using
quantification calibrators for four gene transcripts
(CK-19, EpCAM, MUC1, Her2) containing a known
number of copies, prepared as described in Methods.
These were synthesized as described and aliquoted
to eliminate experimental variation. The basis for
MCF-7 Mean tumor cells recovered 0.00575 0.0065 0.00975 0.01475 0.01575 0.01875 0.02571 0.034125 0.137 adopting this method was to enable comparisons
of CTC analysis as copy number in clinical samples
collected and analyzed at different time points, and
also to allow for the possibility of a low detection limit
such as 1 copy. Figure 5 shows the calibration curve for
Tumor Cells Expected 0.001 0.005 0.01 0.015 0.02 0.025 0.05 0.1 0.5 the four selected transcripts in the range from 10 copy
6
number to 1 copy, with a linearity observed from 10
6
copies to 10 copies with correlation coefficient close
28 Journal of Cancer Metastasis and Treatment ¦ Volume 3 ¦ February 23, 2017