Page 35 - Read Online
P. 35
Page 22 Soren et al. Cancer Drug Resist 2020;3:18-25 I http://dx.doi.org/10.20517/cdr.2019.106
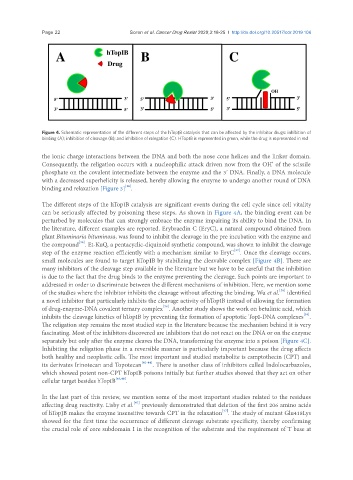
Figure 4. Schematic representation of the different steps of the hTopIB catalysis that can be affected by the inhibitor drugs: inhibition of
binding (A); inhibition of cleavage (B); and inhibition of relegation (C). HTopIB is represented in green, while the drug is represented in red
the ionic charge interactions between the DNA and both the nose cone helices and the linker domain.
Consequently, the religation occurs with a nucleophilic attack driven now from the OH’ of the scissile
phosphate on the covalent intermediate between the enzyme and the 3’ DNA. Finally, a DNA molecule
with a decreased superhelicity is released, hereby allowing the enzyme to undergo another round of DNA
[16]
binding and relaxation [Figure 3] .
The different steps of the hTopIB catalysis are significant events during the cell cycle since cell vitality
can be seriously affected by poisoning these steps. As shown in Figure 4A, the binding event can be
perturbed by molecules that can strongly embrace the enzyme impairing its ability to bind the DNA. In
the literature, different examples are reported. Erybraedin C (EryC), a natural compound obtained from
plant Bituminaria bituminosa, was found to inhibit the cleavage in the pre incubation with the enzyme and
[36]
the compound . Et-KuQ, a pentacyclic-diquinoid synthetic compound, was shown to inhibit the cleavage
[37]
step of the enzyme reaction efficiently with a mechanism similar to EryC . Once the cleavage occurs,
small molecules are found to target hTopIB by stabilizing the cleavable complex [Figure 4B]. There are
many inhibitors of the cleavage step available in the literature but we have to be careful that the inhibition
is due to the fact that the drug binds to the enzyme preventing the cleavage. Such points are important to
addressed in order to discriminate between the different mechanisms of inhibition. Here, we mention some
[38]
of the studies where the inhibitor inhibits the cleavage without affecting the binding. Wu et al. identified
a novel inhibitor that particularly inhibits the cleavage activity of hTopIB instead of allowing the formation
[38]
of drug-enzyme-DNA covalent ternary complex . Another study shows the work on betulinic acid, which
inhibits the cleavage kinetics of hTopIB by preventing the formation of apoptotic TopI-DNA complexes .
[39]
The religation step remains the most studied step in the literature because the mechanism behind it is very
fascinating. Most of the inhibitors discovered are inhibitors that do not react on the DNA or on the enzyme
separately but only after the enzyme cleaves the DNA, transforming the enzyme into a poison [Figure 4C].
Inhibiting the religation phase in a reversible manner is particularly important because the drug affects
both healthy and neoplastic cells. The most important and studied metabolite is camptothecin (CPT) and
its derivates Irinotecan and Topotecan [40-44] . There is another class of inhibitors called Indolocarbazoles,
which showed potent non-CPT hTopIB poisons initially but further studies showed that they act on other
cellular target besides hTopIB [45,46] .
In the last part of this review, we mention some of the most important studies related to the residues
[47]
affecting drug reactivity. Lisby et al. previously demonstrated that deletion of the first 206 amino acids
[47]
of hTopIB makes the enzyme insensitive towards CPT in the relaxation . The study of mutant Glu418Lys
showed for the first time the occurrence of different cleavage substrate specificity, thereby confirming
the crucial role of core subdomain I in the recognition of the substrate and the requirement of T base at