Page 33 - Read Online
P. 33
Page 20 Soren et al. Cancer Drug Resist 2020;3:18-25 I http://dx.doi.org/10.20517/cdr.2019.106
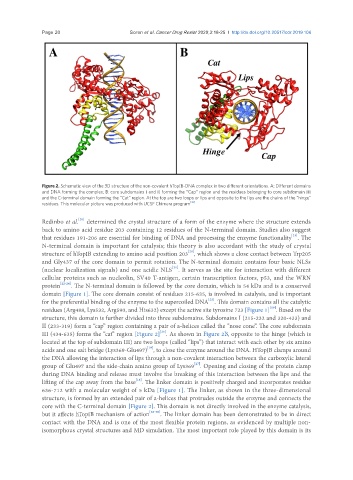
Figure 2. Schematic view of the 3D structure of the non-covalent hTopIB-DNA complex in two different orientations. A: Different domains
and DNA forming the complex; B: core subdomains I and II forming the “Cap” region and the residues belonging to core subdomain III
and the C-terminal domain forming the “Cat” region. At the top are two loops or lips and opposite to the lips are the chains of the “hinge”
residues. This molecular picture was produced with UCSF Chimera program [35]
[20]
Redinbo et al. determined the crystal structure of a form of the enzyme where the structure extends
back to amino acid residue 203 containing 12 residues of the N-terminal domain. Studies also suggest
[21]
that residues 191-206 are essential for binding of DNA and processing the enzyme functionality . The
N-terminal domain is important for catalysis; this theory is also accordant with the study of crystal
structure of hTopIB extending to amino acid position 203 , which shows a close contact between Trp205
[20]
and Gly437 of the core domain to permit rotation. The N-terminal domain contains four basic NLSs
(nuclear localization signals) and one acidic NLS . It serves as the site for interaction with different
[21]
cellular proteins such as nucleolin, SV40 T-antigen, certain transcription factors, p53, and the WRN
protein [22-24] . The N-terminal domain is followed by the core domain, which is 54 kDa and is a conserved
domain [Figure 1]. The core domain consist of residues 215-635, is involved in catalysis, and is important
[25]
for the preferential binding of the enzyme to the supercoiled DNA . This domain contains all the catalytic
[19]
residues (Arg488, Lys532, Arg590, and His632) except the active site tyrosine 723 [Figure 1] . Based on the
structure, this domain is further divided into three subdomains. Subdomains I (215-232 and 320-433) and
II (233-319) form a “cap” region containing a pair of a-helices called the “nose cone”. The core subdomain
III (434-635) forms the “cat” region [Figure 2] . As shown in Figure 2B, opposite to the hinge (which is
[26]
located at the top of subdomain III) are two loops (called “lips”) that interact with each other by six amino
[19]
acids and one salt bridge (Lys369-Glu497) , to close the enzyme around the DNA. HTopIB clamps around
the DNA allowing the interaction of lips through a non-covalent interaction between the carboxylic lateral
group of Glu497 and the side-chain amino group of Lys369 . Opening and closing of the protein clamp
[27]
during DNA binding and release must involve the breaking of this interaction between the lips and the
lifting of the cap away from the base . The linker domain is positively charged and incorporates residue
[16]
636-712 with a molecular weight of 5 kDa [Figure 1]. The linker, as shown in the three-dimensional
structure, is formed by an extended pair of a-helices that protrudes outside the enzyme and connects the
core with the C-terminal domain [Figure 2]. This domain is not directly involved in the enzyme catalysis,
but it affects hTopIB mechanism of action [28-30] . The linker domain has been demonstrated to be in direct
contact with the DNA and is one of the most flexible protein regions, as evidenced by multiple non-
isomorphous crystal structures and MD simulation. The most important role played by this domain is its