Page 34 - Read Online
P. 34
Soren et al. Cancer Drug Resist 2020;3:18-25 I http://dx.doi.org/10.20517/cdr.2019.106 Page 21
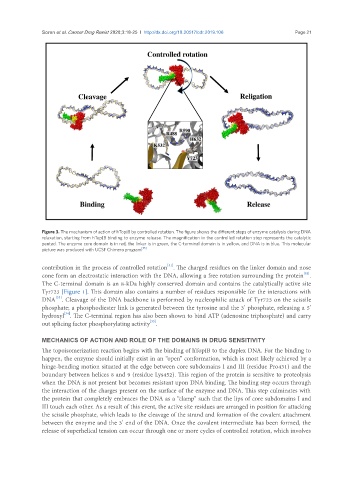
Figure 3. The mechanism of action of hTopIB by controlled rotation. The figure shows the different steps of enzyme catalysis during DNA
relaxation, starting from hTopIB binding to enzyme release. The magnification in the controlled rotation step represents the catalytic
pentad. The enzyme core domain is in red, the linker is in green, the C-terminal domain is in yellow, and DNA is in blue. This molecular
picture was produced with UCSF Chimera program [35]
[31]
contribution in the process of controlled rotation . The charged residues on the linker domain and nose
[32]
cone form an electrostatic interaction with the DNA, allowing a free rotation surrounding the protein .
The C-terminal domain is an 8-kDa highly conserved domain and contains the catalytically active site
Tyr723 [Figure 1]. This domain also contains a number of residues responsible for the interactions with
[33]
DNA . Cleavage of the DNA backbone is performed by nucleophilic attack of Tyr723 on the scissile
phosphate; a phosphodiester link is generated between the tyrosine and the 3’ phosphate, releasing a 5’
[34]
hydroxyl . The C-terminal region has also been shown to bind ATP (adenosine triphosphate) and carry
[33]
out splicing factor phosphorylating activity .
MECHANICS OF ACTION AND ROLE OF THE DOMAINS IN DRUG SENSITIVITY
The topoisomerization reaction begins with the binding of hTopIB to the duplex DNA. For the binding to
happen, the enzyme should initially exist in an “open” conformation, which is most likely achieved by a
hinge-bending motion situated at the edge between core subdomains I and III (residue Pro431) and the
boundary between helices 8 and 9 (residue Lys452). This region of the protein is sensitive to proteolysis
when the DNA is not present but becomes resistant upon DNA binding. The binding step occurs through
the interaction of the charges present on the surface of the enzyme and DNA. This step culminates with
the protein that completely embraces the DNA as a “clamp” such that the lips of core subdomains I and
III touch each other. As a result of this event, the active site residues are arranged in position for attacking
the scissile phosphate, which leads to the cleavage of the strand and formation of the covalent attachment
between the enzyme and the 3’ end of the DNA. Once the covalent intermediate has been formed, the
release of superhelical tension can occur through one or more cycles of controlled rotation, which involves