Page 17 - Read Online
P. 17
Page 8 of 15 Chen et al. Cancer Drug Resist 2024;7:9 https://dx.doi.org/10.20517/cdr.2023.151
+
+
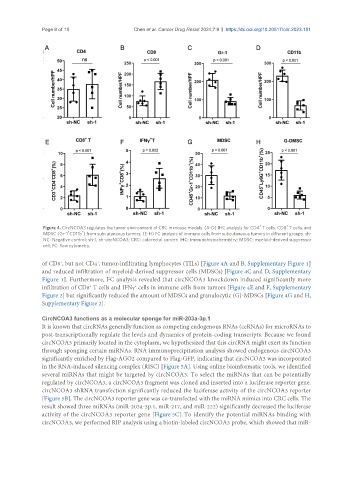
Figure 4. CircNCOA3 regulates the tumor environment of CRC in mouse models. (A-D) IHC analysis for CD4 T cells, CD8 T cells, and
+
+
MDSC (Gr-1 CD11b ) from subcutaneous tumors; (E-H) FC analysis of immune cells from subcutaneous tumors in different groups. sh-
NC: Negative control; sh-1, sh-circNCOA3; CRC: colorectal cancer; IHC: immunohistochemistry; MDSC: myeloid-derived suppressor
cell; FC: flow cytometry.
+
of CD8 , but not CD4 , tumor-infiltrating lymphocytes (TILs) [Figure 4A and B, Supplementary Figure 1]
+
and reduced infiltration of myeloid-derived suppressor cells (MDSCs) [Figure 4C and D, Supplementary
Figure 1]. Furthermore, FC analysis revealed that circNCOA3 knockdown induced significantly more
infiltration of CD8 T cells and IFNγ cells in immune cells from tumors [Figure 4E and F, Supplementary
+
+
Figure 2] but significantly reduced the amount of MDSCs and granulocytic (G)-MDSCs [Figure 4G and H,
Supplementary Figure 2].
CircNCOA3 functions as a molecular sponge for miR-203a-3p.1
It is known that circRNAs generally function as competing endogenous RNAs (ceRNAs) for microRNAs to
post-transcriptionally regulate the levels and dynamics of protein-coding transcripts. Because we found
circNCOA3 primarily located in the cytoplasm, we hypothesized that this circRNA might exert its function
through sponging certain miRNAs. RNA immunoprecipitation analysis showed endogenous circNCOA3
significantly enriched by Flag-AGO2 compared to Flag-GFP, indicating that circNCOA3 was incorporated
in the RNA-induced silencing complex (RISC) [Figure 5A]. Using online bioinformatic tools, we identified
several miRNAs that might be targeted by circNCOA3. To select the miRNAs that can be potentially
regulated by circNCOA3, a circNCOA3 fragment was cloned and inserted into a luciferase reporter gene.
circNCOA3 shRNA transfection significantly reduced the luciferase activity of the circNCOA3 reporter
[Figure 5B]. The circNCOA3 reporter gene was co-transfected with the miRNA mimics into CRC cells. The
result showed three miRNAs (miR-203a-3p.1, miR-217, and miR-222) significantly decreased the luciferase
activity of the circNCOA3 reporter gene [Figure 5C]. To identify the potential miRNAs binding with
circNCOA3, we performed RIP analysis using a biotin-labeled circNCOA3 probe, which showed that miR-