Page 15 - Read Online
P. 15
Page 6 of 15 Chen et al. Cancer Drug Resist 2024;7:9 https://dx.doi.org/10.20517/cdr.2023.151
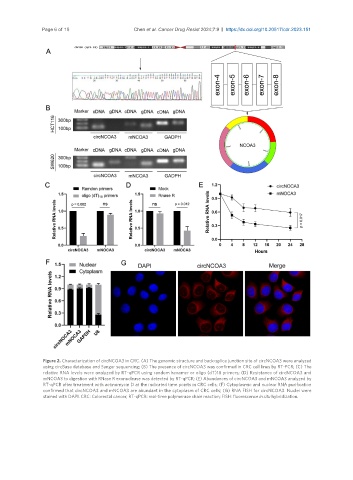
Figure 2. Characterization of circNCOA3 in CRC. (A) The genomic structure and back-splice junction site of circNCOA3 were analyzed
using circBase database and Sanger sequencing; (B) The presence of circNCOA3 was confirmed in CRC cell lines by RT-PCR; (C) The
relative RNA levels were analyzed by RT-qPCR using random hexamer or oligo (dT)18 primers; (D) Resistance of circNCOA3 and
mNCOA3 to digestion with RNase R exonuclease was detected by RT-qPCR; (E) Abundances of circNCOA3 and mNCOA3 analyzed by
RT-qPCR after treatment with actinomycin D at the indicated time points in CRC cells; (F) Cytoplasmic and nuclear RNA purification
confirmed that circNCOA3 and mNCOA3 are abundant in the cytoplasm of CRC cells; (G) RNA FISH for circNCOA3. Nuclei were
stained with DAPI. CRC: Colorectal cancer; RT-qPCR: real-time polymerase chain reaction; FISH: fluorescence in situ hybridization.