Page 13 - Read Online
P. 13
Skoreński et al. Rare Dis Orphan Drugs J 2023;2:6 https://dx.doi.org/10.20517/rdodj.2022.21 Page 9 of 23
[44-46]
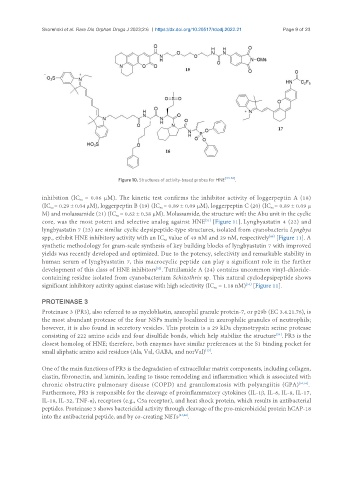
Figure 10. Structures of activity-based probes for HNE .
inhibition (IC = 0.06 µM). The kinetic test confirms the inhibitor activity of loggerpeptin A (18)
50
(IC = 0.29 ± 0.04 µM), loggerpeptin B (19) (IC = 0.89 ± 0.09 µM), loggerpeptin C (20) (IC = 0.89 ± 0.09 µ
50
50
50
M) and molassamide (21) (IC = 0.62 ± 0.38 µM). Molassamide, the structure with the Abu unit in the cyclic
50
core, was the most potent and selective analog against HNE [Figure 11]. Lyngbyastatin 4 (22) and
[51]
lyngbyastatin 7 (23) are similar cyclic depsipeptide-type structures, isolated from cyanobacteria Lyngbya
spp., exhibit HNE inhibitory activity with an IC value of 49 nM and 29 nM, respectively [Figure 11]. A
[48]
50
synthetic methodology for gram-scale synthesis of key building blocks of lyngbyastatin 7 with improved
yields was recently developed and optimized. Due to the potency, selectivity and remarkable stability in
human serum of lyngbyastatin 7, this macrocyclic peptide can play a significant role in the further
development of this class of HNE inhibitors . Tutuilamide A (24) contains uncommon vinyl-chloride-
[52]
containing residue isolated from cyanobacterium Schizothrix sp. This natural cyclodepsipeptide shows
[53]
significant inhibitory activity against elastase with high selectivity (IC = 1.18 nM) [Figure 11].
50
PROTEINASE 3
Proteinase 3 (PR3), also referred to as myeloblastin, azurophil granule protein-7, or p29b (EC 3.4.21.76), is
the most abundant protease of the four NSPs mainly localized in azurophilic granules of neutrophils;
however, it is also found in secretory vesicles. This protein is a 29 kDa chymotrypsin serine protease
[54]
consisting of 222 amino acids and four disulfide bonds, which help stabilize the structure . PR3 is the
closest homolog of HNE; therefore, both enzymes have similar preferences at the S1 binding pocket for
small aliphatic amino acid residues (Ala, Val, GABA, and norVal) .
[15]
One of the main functions of PR3 is the degradation of extracellular matrix components, including collagen,
elastin, fibronectin, and laminin, leading to tissue remodeling and inflammation which is associated with
chronic obstructive pulmonary disease (COPD) and granulomatosis with polyangiitis (GPA) [54,55] .
Furthermore, PR3 is responsible for the cleavage of proinflammatory cytokines (IL-1β, IL-6, IL-8, IL-17,
IL-18, IL-32, TNF-α), receptors (e.g., C5a receptor), and heat shock protein, which results in antibacterial
peptides. Proteinase 3 shows bactericidal activity through cleavage of the pro-microbicidal protein hCAP-18
into the antibacterial peptide, and by co-creating NETs [54,56] .