Page 18 - Read Online
P. 18
Page 4 of 14 Ricci et al. Vessel Plus 2021;5:31 https://dx.doi.org/10.20517/2574-1209.2021.28
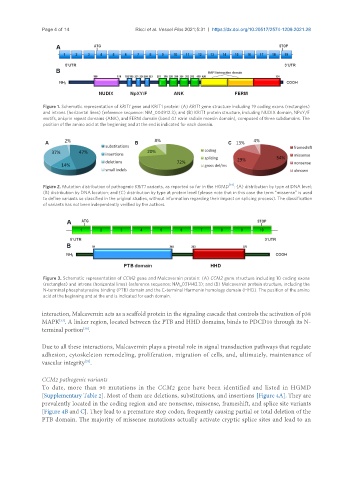
Figure 1. Schematic representation of KRIT1 gene and KRIT1 protein: (A) KRIT1 gene structure including 19 coding exons (rectangles)
and introns (horizontal lines) (reference sequence: NM_004912.3); and (B) KRIT1 protein structure, including NUDIX domain, NPxY/F
motifs, ankyrin repeat domains (ANK), and FERM domain (band 4.1 ezrin radixin moesin domain), composed of three subdomains. The
position of the amino acid at the beginning and at the end is indicated for each domain.
Figure 2. Mutation distribution of pathogenic KRIT1 variants, as reported so far in the HGMD [14] : (A) distribution by type at DNA level;
(B) distribution by DNA location; and (C) distribution by type at protein level (please note that in this case the term “missense” is used
to define variants so classified in the original studies, without information regarding their impact on splicing process). The classification
of variants has not been independently verified by the authors.
Figure 3. Schematic representation of CCM2 gene and Malcavernin protein: (A) CCM2 gene structure including 10 coding exons
(rectangles) and introns (horizontal lines) (reference sequence: NM_031443.3); and (B) Malcavernin protein structure, including the
N-terminal phosphotyrosine binding (PTB) domain and the C-terminal Harmonin homology domain (HHD). The position of the amino
acid at the beginning and at the end is indicated for each domain.
interaction, Malcavernin acts as a scaffold protein in the signaling cascade that controls the activation of p38
MAPK . A linker region, located between the PTB and HHD domains, binds to PDCD10 through its N-
[33]
[38]
terminal portion .
Due to all these interactions, Malcavernin plays a pivotal role in signal transduction pathways that regulate
adhesion, cytoskeleton remodeling, proliferation, migration of cells, and, ultimately, maintenance of
vascular integrity .
[33]
CCM2 pathogenic variants
To date, more than 90 mutations in the CCM2 gene have been identified and listed in HGMD
[Supplementary Table 2]. Most of them are deletions, substitutions, and insertions [Figure 4A]. They are
prevalently located in the coding region and are nonsense, missense, frameshift, and splice site variants
[Figure 4B and C]. They lead to a premature stop codon, frequently causing partial or total deletion of the
PTB domain. The majority of missense mutations actually activate cryptic splice sites and lead to an