Page 79 - Read Online
P. 79
Page 10 of 16 Renzi et al. Microbiome Res Rep 2024;3:2 https://dx.doi.org/10.20517/mrr.2023.27
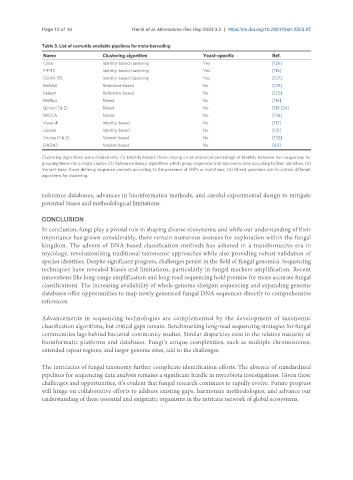
Table 3. List of currently available pipelines for meta-barcoding
Name Clustering algorithm Yeast-specific Ref.
Clotu Identity-based clustering Yes [126]
PIPITS Identity-based clustering Yes [116]
CloVR-ITS Identity-based clustering Yes [127]
BioMaS Reference-based No [129]
Kraken Reference-based No [123]
Mothur Mixed No [115]
Qiime (1 & 2) Mixed No [119,124]
MICCA Mixed No [128]
Vsearch Identity-based No [117]
Uparse Identity-based No [131]
Unoise (1 & 2) Variant-based No [132]
DADA2 Variant-based No [60]
Clustering algorithms were divided into: (1) Identity-based, those relying on an empirical percentage of identity between two sequences for
grouping them into a single cluster; (2) Reference-based, algorithms which group sequences into taxonomic bins according to their identities; (3)
Variant-base, those defining sequence variants according to the presence of SNPs or mutations; (4) Mixed, pipelines which contain different
algorithms for clustering.
reference databases, advances in bioinformatics methods, and careful experimental design to mitigate
potential biases and methodological limitations.
CONCLUSION
In conclusion, fungi play a pivotal role in shaping diverse ecosystems, and while our understanding of their
importance has grown considerably, there remain numerous avenues for exploration within the fungal
kingdom. The advent of DNA-based classification methods has ushered in a transformative era in
mycology, revolutionizing traditional taxonomic approaches while also providing robust validation of
species identities. Despite significant progress, challenges persist in the field of fungal genomics. Sequencing
techniques have revealed biases and limitations, particularly in fungal markers amplification. Recent
innovations like long-range amplification and long-read sequencing hold promise for more accurate fungal
classifications. The increasing availability of whole-genome shotgun sequencing and expanding genome
databases offer opportunities to map newly generated fungal DNA sequences directly to comprehensive
references.
Advancements in sequencing technologies are complemented by the development of taxonomic
classification algorithms, but critical gaps remain. Benchmarking long-read sequencing strategies for fungal
communities lags behind bacterial community studies. Similar disparities exist in the relative maturity of
bioinformatic platforms and databases. Fungi’s unique complexities, such as multiple chromosomes,
extended repeat regions, and larger genome sizes, add to the challenges.
The intricacies of fungal taxonomy further complicate identification efforts. The absence of standardized
pipelines for sequencing data analysis remains a significant hurdle in mycobiota investigations. Given these
challenges and opportunities, it’s evident that fungal research continues to rapidly evolve. Future progress
will hinge on collaborative efforts to address existing gaps, harmonize methodologies, and advance our
understanding of these essential and enigmatic organisms in the intricate network of global ecosystems.