Page 76 - Read Online
P. 76
Renzi et al. Microbiome Res Rep 2024;3:2 https://dx.doi.org/10.20517/mrr.2023.27 Page 7 of 16
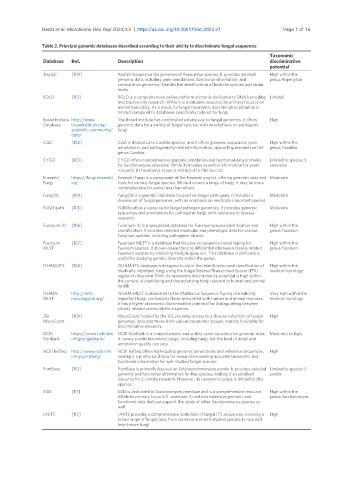
Table 2. Principal genomic databases described according to their ability to discriminate fungal sequences
Taxonomic
Database Ref. Description discriminative
potential
AspGD [100] AspGD focuses on the genomes of Aspergillus species. It provides detailed High within the
genomic data, including gene annotations, functional information, and genus Aspergillus
comparative genomics. Enables the identification of both the species and strain
levels.
BOLD [101] BOLD is a comprehensive online platform primarily dedicated to DNA barcoding Limited
and biodiversity research. While it is a valuable resource, its primary focus is on
animal barcoding. As a result, its fungal taxonomic discriminative potential is
limited compared to databases specifically tailored for fungi.
Broad Insitute http://www. The Broad Institute has contributed extensively to fungal genomics. It offers High
Database broadinstitute.org/ genomic data for a variety of fungal species, with an emphasis on pathogenic
scientific-community/ fungi.
data/
CGD [102] CGD is dedicated to Candida species, and it offers genomic sequences, gene High within the
annotations, and pathogenicity-related information, supporting research on the genus Candida
genus Candida.
CYGD [103] CYGD offers comprehensive genome annotation and functional data primarily Limited to species S.
for Saccharomyces cerevisiae. While it provides essential information for yeast cerevisiae
research, its taxonomic scope is restricted to this species.
Ensembl https://fungi.ensembl. Ensembl Fungi is a component of the Ensembl project, offering genomic data and Moderate
Fungi org tools for various fungal species. While it covers a range of fungi, it may be more
comprehensive for some taxa than others.
FungiDB [104] FungiDB is a genomic database focused on fungal pathogens. It includes a Moderate
diverse set of fungal genomes, with an emphasis on medically important species.
FUNGIpath [105] FUNGIpath is a resource for fungal pathogen genomics. It provides genomic Moderate
sequences and annotations for pathogenic fungi, with relevance to disease
research.
Fusarium-ID [106] Fusarium-ID is a specialized database for Fusarium species identification and High within the
classification. It provides detailed molecular and phenotypic data for various genus Fusarium
Fusarium species, including pathogenic strains.
Fusarium [107] Fusarium MLST is a database that focuses on sequence-based typing for High within the
MLST Fusarium species. It allows researchers to differentiate between closely related genus Fusarium
Fusarium isolates by analyzing multiple gene loci. This database is particularly
useful for studying genetic diversity within the genus.
ISHAM-ITS [108] ISHAM-ITS database is designed to aid in the identification and classification of High within the
medically important fungi using the fungal Internal Transcribed Spacer (ITS) medical mycology
region of ribosomal DNA. Its taxonomic discriminative potential is high within
the context of identifying and characterizing fungi relevant to human and animal
health.
ISHAM- http://mlst. ISHAM-MLST is dedicated to the Multilocus Sequence Typing of medically Very high within the
MLST mycologylab.org/ important fungi, particularly those associated with human and animal mycoses. medical mycology
It has a higher taxonomic discriminative potential for distinguishing between
closely related strains within a species.
JGI [109] MycoCosm, hosted by the JGI, provides access to a diverse collection of fungal High
MycoCosm genomes, including those from various taxonomic groups, making it suitable for
discriminative research.
NCBI https://www.ncbi.nlm. NCBI GenBank is a comprehensive and widely used repository for genomic data. Moderate to high
GenBank nih.gov/genbank/ It covers a wide taxonomic range, including fungi, but the level of detail and
annotation quality can vary.
NCBI RefSeq http://www.ncbi.nlm. NCBI RefSeq offers high-quality genomic annotations and reference sequences, High
nih.gov/refseq/ making it a preferred choice for researchers seeking accurate taxonomic and
functional information for well-studied fungal species.
PomBase [110] PomBase is primarily focused on Schizosaccharomyces pombe. It provides detailed Limited to species S.
genomic and functional information for this species, making it an excellent pombe
resource for S. pombe research. However, its taxonomic scope is limited to this
species.
SGD [111] SGD is dedicated to Saccharomyces cerevisiae and is a comprehensive resource. High within the
While its primary focus is S. cerevisiae, it contains extensive genomic and genus Saccharomyces
functional data that can support the study of other Saccharomyces species as
well.
UNITE [112] UNITE provides a comprehensive collection of fungal ITS sequences, covering a High
broad range of fungal taxa, from common and well-studied species to rare and
less-known fungi.