Page 70 - Read Online
P. 70
Ghaseminejad et al. J Transl Genet Genom 2022;6:111-25 https://dx.doi.org/10.20517/jtgg.2021.49 Page 115
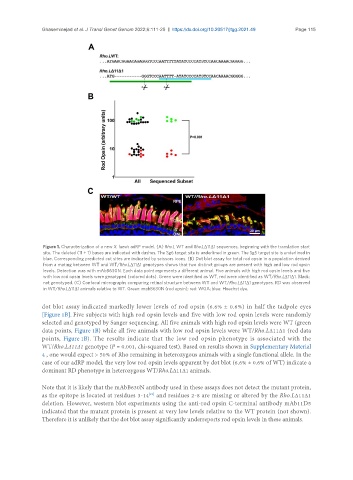
Figure 1. Characterization of a new X. laevis adRP model. (A) Rho.L WT and Rho.LΔ11Δ1 sequences, beginning with the translation start
site. The deleted (11 + 1) bases are indicated with dashes. The Sg6 target site is underlined in green. The Sg5 target site is underlined in
blue. Corresponding predicted cut sites are indicated by scissors icons. (B) Dot blot assay for total rod opsin in a population derived
from a mating between WT and WT/Rho.LΔ11Δ1 genotypes shows that two distinct groups are present with high and low rod opsin
levels. Detection was with mAbB630N. Each data point represents a different animal. Five animals with high rod opsin levels and five
with low rod opsin levels were genotyped (colored dots). Green were identified as WT, red were identified as WT/Rho.LΔ11Δ1. Black:
not genotyped. (C) Confocal micrographs comparing retinal structure between WT and WT/Rho.LΔ11Δ1 genotypes. RD was observed
in WT/Rho.LΔ11Δ1 animals relative to WT. Green: mabB630N (rod opsin); red: WGA; blue: Hoechst dye.
dot blot assay indicated markedly lower levels of rod opsin (6.6% ± 0.6%) in half the tadpole eyes
[Figure 1B]. Five subjects with high rod opsin levels and five with low rod opsin levels were randomly
selected and genotyped by Sanger sequencing. All five animals with high rod opsin levels were WT (green
data points, Figure 1B) while all five animals with low rod opsin levels were WT/Rho.LΔ11Δ1 (red data
points, Figure 1B). The results indicate that the low rod opsin phenotype is associated with the
WT/Rho.LΔ11Δ1 genotype (P = 0.001, chi-squared test). Based on results shown in Supplementary Material
4 , one would expect > 50% of Rho remaining in heterozygous animals with a single functional allele. In the
case of our adRP model, the very low rod opsin levels apparent by dot blot (6.6% ± 0.6% of WT) indicate a
dominant RD phenotype in heterozygous WT/Rho.LΔ11Δ1 animals.
Note that it is likely that the mAbB630N antibody used in these assays does not detect the mutant protein,
[21]
as the epitope is located at residues 3-14 and residues 2-8 are missing or altered by the Rho.LΔ11Δ1
deletion. However, western blot experiments using the anti-rod opsin C-terminal antibody mAb11D5
indicated that the mutant protein is present at very low levels relative to the WT protein (not shown).
Therefore it is unlikely that the dot blot assay significantly underreports rod opsin levels in these animals.