Page 47 - Read Online
P. 47
Page 119 Waller et al. J Transl Genet Genom 2021;5:112-23 I http://dx.doi.org/10.20517/jtgg.2021.09
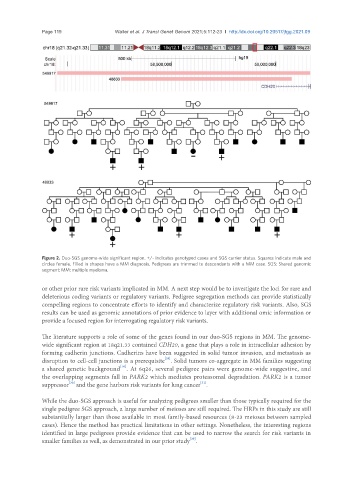
Figure 2. Duo-SGS genome-wide significant region. +/- indicates genotyped cases and SGS carrier status. Squares indicate male and
circles female. Filled in shapes have a MM diagnosis. Pedigrees are trimmed to descendants with a MM case. SGS: Shared genomic
segment; MM: multiple myeloma.
or other prior rare risk variants implicated in MM. A next step would be to investigate the loci for rare and
deleterious coding variants or regulatory variants. Pedigree segregation methods can provide statistically
compelling regions to concentrate efforts to identify and characterize regulatory risk variants. Also, SGS
results can be used as genomic annotations of prior evidence to layer with additional omic information or
provide a focused region for interrogating regulatory risk variants.
The literature supports a role of some of the genes found in our duo-SGS regions in MM. The genome-
wide significant region at 18q21.33 contained CDH20, a gene that plays a role in intracellular adhesion by
forming cadherin junctions. Cadherins have been suggested in solid tumor invasion, and metastasis as
[29]
disruption to cell-cell junctions is a prerequisite . Solid tumors co-aggregate in MM families suggesting
[10]
a shared genetic background . At 6q26, several pedigree pairs were genome-wide suggestive, and
the overlapping segments fall in PARK2 which mediates proteasomal degradation. PARK2 is a tumor
[31]
[30]
suppressor and the gene harbors risk variants for lung cancer .
While the duo-SGS approach is useful for analyzing pedigrees smaller than those typically required for the
single pedigree SGS approach, a large number of meioses are still required. The HRPs in this study are still
substantially larger than those available in most family-based resources (8-23 meioses between sampled
cases). Hence the method has practical limitations in other settings. Nonetheless, the interesting regions
identified in large pedigrees provide evidence that can be used to narrow the search for risk variants in
[20]
smaller families as well, as demonstrated in our prior study .