Page 33 - Read Online
P. 33
Zhang et al. J Transl Genet Genom 2018;2:18. I https://doi.org/10.20517/jtgg.2018.22 Page 9 of 11
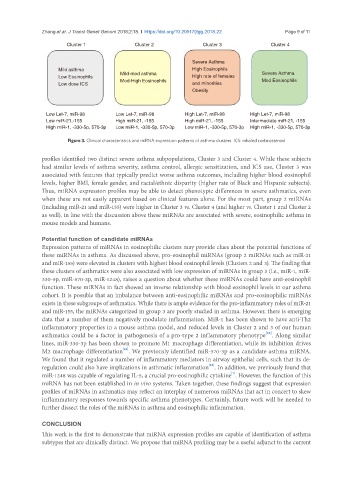
Cluster 1 Cluster 2 Cluster 3 Cluster 4
Severe Asthma
Mild asthma High Eosinophils
Low Eosinophils Mild-mod asthma High rate of females Severe Asthma
Low dose ICS Mod-High Eosinophils and minorities Mod Eosinophils
Obesity
Low Let-7, miR-98 Low Let-7, miR-98 High Let-7, miR-98 High Let-7, miR-98
Low miR-21,-155 High miR-21, -155 High miR-21, -155 Intermediate miR-21, -155
High miR-1, -330-5p, 570-3p Low miR-1, -330-5p, 570-3p Low miR-1, -330-5p, 570-3p High miR-1, -330-5p, 570-3p
Figure 3. Clinical characteristics and miRNA expression patterns of asthma clusters. ICS: inhaled corticosteroid
profiles identified two distinct severe asthma subpopulations, Cluster 3 and Cluster 4. While these subjects
had similar levels of asthma severity, asthma control, allergic sensitization, and ICS use, Cluster 3 was
associated with features that typically predict worse asthma outcomes, including higher blood eosinophil
levels, higher BMI, female gender, and racial/ethnic disparity (higher rate of Black and Hispanic subjects).
Thus, miRNA expression profiles may be able to detect phenotypic differences in severe asthmatics, even
when these are not easily apparent based on clinical features alone. For the most part, group 2 miRNAs
(including miR-21 and miR-155) were higher in Cluster 3 vs. Cluster 4 (and higher vs. Cluster 1 and Cluster 2
as well), in line with the discussion above these miRNAs are associated with severe, eosinophilic asthma in
mouse models and humans.
Potential function of candidate miRNAs
Expression patterns of miRNAs in eosinophilic clusters may provide clues about the potential functions of
these miRNAs in asthma. As discussed above, pro-eosinophil miRNAs (group 2 miRNAs such as miR-21
and miR-155) were elevated in clusters with highest blood eosinophil levels (Clusters 2 and 3). The finding that
these clusters of asthmatics were also associated with low expression of miRNAs in group 3 (i.e., miR-1, miR-
330-5p, miR-570-3p, miR-1248), raises a question about whether these miRNAs could have anti-eosinophil
function. These miRNAs in fact showed an inverse relationship with blood eosinophil levels in our asthma
cohort. It is possible that an imbalance between anti-eosinophilic miRNAs and pro-eosinophilic miRNAs
exists in these subgroups of asthmatics. While there is ample evidence for the pro-inflammatory roles of miR-21
and miR-155, the miRNAs categorized in group 3 are poorly studied in asthma. However, there is emerging
data that a number of them negatively modulate inflammation. MiR-1 has been shown to have anti-Th2
inflammatory properties in a mouse asthma model, and reduced levels in Cluster 2 and 3 of our human
[20]
asthmatics could be a factor in pathogenesis of a pro-type 2 inflammatory phenotype . Along similar
lines, miR-330-3p has been shown to promote M1 macrophage differentiation, while its inhibition drives
[21]
M2 macrophage differentiation . We previously identified miR-570-3p as a candidate asthma miRNA.
We found that it regulated a number of inflammatory mediators in airway epithelial cells, such that its de-
[22]
regulation could also have implications in asthmatic inflammation . In addition, we previously found that
[7]
miR-1248 was capable of regulating IL-5, a crucial pro-eosinophilic cytokine . However, the function of this
miRNA has not been established in in vivo systems. Taken together, these findings suggest that expression
profiles of miRNAs in asthmatics may reflect an interplay of numerous miRNAs that act in concert to skew
inflammatory responses towards specific asthma phenotypes. Certainly, future work will be needed to
further dissect the roles of the miRNAs in asthma and eosinophilic inflammation.
CONCLUSION
This work is the first to demonstrate that miRNA expression profiles are capable of identification of asthma
subtypes that are clinically distinct. We propose that miRNA profiling may be a useful adjunct to the current