Page 25 - Read Online
P. 25
Przanowski et al. J Transl Genet Genom 2018;2:2 I http://dx.doi.org/10.20517/jtgg.2017.03 Page 5 of 15
Biological systems XCIFs screens
Xi-Gfp [32,33,40]
Xi-Luciferase [41]
Ch17-TetO-Xist-Mylc2b-Gfp [42]
Xi-Mecp2-Hygromycin [34] Genetic Proteomic
Ch6-TetO-Xist survival [43] screens screens
RNAi library
Gene-trap (shRNA or siRNA) Chemical library
Number of
screens performed:
Number of XCIFs
found with screen:
Number
of validated XCIFs:
Total number
of validated XCIFs:
Validation methods
qRT-PCR for X-linked genes [40,49] ; RNA-FISH for X-linked genes [32,42,47] ;
Xi-Luciferase [34,41] ; Xi-Gfp [33,48] , Immunostaining for H4Kac [43] ;
Immunostaining for H3K27me3 [47,48]
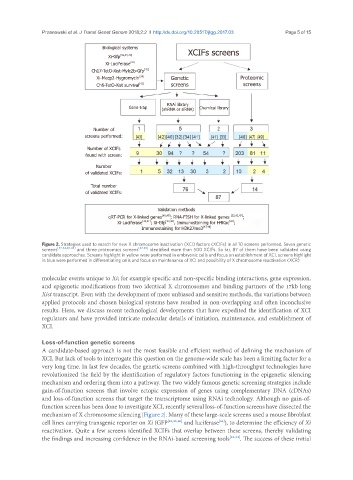
Figure 2. Strategies used to search for new X chromosome inactivation (XCI) factors (XCIFs) in all 10 screens performed. Seven genetic
screens [32-34,40-43] and three proteomics screens [47-49] identified more than 500 XCIFs. So far, 87 of them have been validated using
candidate approaches. Screens highlight in yellow were performed in embryonic cells and focus on establishment of XCI, screens highlight
in blue were performed in differentiating cells and focus on maintenance of XCI and possibility of X chromosome reactivation (XCR)
molecular events unique to Xi; for example specific and non-specific binding interactions, gene expression,
and epigenetic modifications from two identical X chromosomes and binding partners of the 17kb long
Xist transcript. Even with the development of more unbiased and sensitive methods, the variations between
applied protocols and chosen biological systems have resulted in non-overlapping and often inconclusive
results. Here, we discuss recent technological developments that have expedited the identification of XCI
regulators and have provided intricate molecular details of initiation, maintenance, and establishment of
XCI.
Loss-of-function genetic screens
A candidate-based approach is not the most feasible and efficient method of defining the mechanism of
XCI. But lack of tools to interrogate this question on the genome-wide scale has been a limiting factor for a
very long time. In last few decades, the genetic screens combined with high-throughput technologies have
revolutionized the field by the identification of regulatory factors functioning in the epigenetic silencing
mechanism and ordering them into a pathway. The two widely famous genetic screening strategies include
gain-of-function screens that involve ectopic expression of genes using complementary DNA (cDNAs)
and loss-of-function screens that target the transcriptome using RNAi technology. Although no gain-of-
function screen has been done to investigate XCI, recently several loss-of-function screens have dissected the
mechanism of X chromosome silencing [Figure 2]. Many of these large-scale screens used a mouse fibroblast
[34]
cell lines carrying transgenic reporter on Xi (GFP [32,33,40] and luciferase ), to determine the efficiency of Xi
reactivation. Quite a few screens identified XCIFs that overlap between these screens, thereby validating
the findings and increasing confidence in the RNAi-based screening tools [32-34] . The success of these initial