Page 76 - Read Online
P. 76
Ambion, Life Technology), which was used as
spike-in by adding it during the lysis step of miRNAs
extraction.
Results
Detection of ex-miRNAs in cultured medium of
MB cell lines by microarray analysis
Given that some human cancer cells secrete miRNAs
into their extracellular environment and body fl uids, [24-26]
it was hypothesized that MB cell lines may secrete
miRNAs into their spent culture medium. To test this
hypothesis, 3 cell lines representing MB subtypes D341
and D283 (metastasis-related group 3 and group 4 MB
subtypes) and DAOY (sonic hedgehog-related) were
[27]
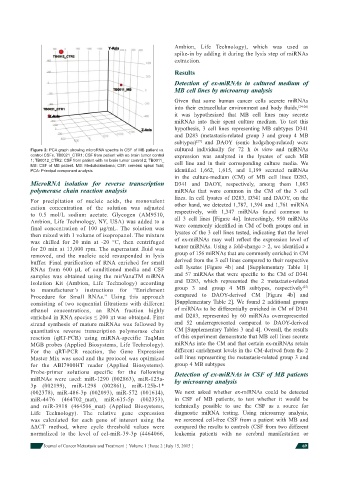
Figure 2: PCA graph showing microRNA spectra in CSF of MB patient vs. cultured individually for 72 h in vitro and miRNAs
control CSFs. TB0021_CTR1: CSF from patient with no brain tumor control expression was analyzed in the lysates of each MB
1; TB0012_CTR2: CSF from patient with no brain tumor control 2; TB0011_ cell line and in their corresponding culture media. We
MB: CSF of MB patient. MB: Medulloblastoma; CSF: cerebral spinal fl uid;
PCA: Principal component analysis identifi ed 1,662, 1,615, and 1,199 secreted miRNAs
in the culture-medium (CM) of MB cell lines D283,
MicroRNA isolation for reverse transcription D341 and DAOY, respectively, among them 1,083
polymerase chain reaction analysis miRNAs that were common in the CM of the 3 cell
lines. In cell lysates of D283, D341 and DAOY, on the
For precipitation of nucleic acids, the monovalent other hand, we detected 1,787, 1,394 and 1,761 miRNA
cation concentration of the solution was adjusted
to 0.5 mol/L sodium acetate. Glycogen (AM9510, respectively, with 1,347 miRNAs found common to
Ambion, Life Technology, NY, USA) was added to a all 3 cell lines [Figure 4a]. Interestingly, 950 miRNAs
final concentration of 100 μg/mL. The solution was were commonly identifi ed in CM of both groups and in
then mixed with 1 volume of isopropanol. The mixture lysates of the 3 cell lines tested, indicating that the level
was chilled for 20 min at -20 °C, then centrifuged of ex-miRNAs may well refl ect the expression level of
for 20 min at 13,000 rpm. The supernatant fluid was tumor miRNAs. Using a fold-change > 2, we identifi ed a
removed, and the nucleic acid resuspended in lysis group of 156 miRNAs that are commonly enriched in CM
buffer. Final purification of RNA enriched for small derived from the 3 cell lines compared to their respective
RNAs from 600 μL of conditioned media and CSF cell lysates [Figure 4b] and [Supplementary Table 1]
samples was obtained using the mirVanaTM miRNA and 57 miRNAs that were spec ifi c to the CM of D341
Isolation Kit (Ambion, Life Technology) according and D283, which represented the 2 metastasis-related
[27]
to manufacturer’s instructions for “Enrichment group 3 and group 4 MB subtypes, respectively
Procedure for Small RNAs.” Using this approach compared to DAOY-derived CM [Figure 4b] and
consisting of two sequential filtrations with different [Supplementary Table 2]. We found 2 additional groups
ethanol concentrations, an RNA fraction highly of miRNAs to be differentially enriched in CM of D341
enriched in RNA species ≤ 200 nt was obtained. First and D283, represented by 60 miRNAs overrepresented
strand synthesis of mature miRNAs was followed by and 52 underrepresented compared to DAOY-derived
quantitative reverse transcription polymerase chain CM [Supplementary Tables 3 and 4]. Overall, the results
reaction (qRT-PCR) using miRNA-specific TaqMan of this experiment demonstrate that MB cell lines secrete
MGB probes (Applied Biosystems, Life Technology). miRNAs into the CM and that certain ex-miRNAs retain
For the qRT-PCR reaction, the Gene Expression different enrichment levels in the CM-derived from the 2
Master Mix was used and the protocol was optimized cell lines representing the metastasis-related group 3 and
for the ABI7900HT reader (Applied Biosystems). group 4 MB subtypes
Probe-primer solutions specific for the following Detection of ex-miRNAs in CSF of MB patients
miRNAs were used: miR-1290 (002863), miR-125a- by microarray analysis
3p (002199), miR-1298 (002861), miR-125b-1*
(002378), miR-486-3p (002093), miR-572 (001614), We next asked whether ex-miRNAs could be detected
miR-4476 (464702_mat), miR-615-5p (002353), in CSF of MB patients, to test whether it would be
and miR-3918 (464506_mat) (Applied Biosystems, technically possible to use the CSF as a source for
Life Technology). The relative gene expression diagnostic miRNA testing. Using microarray analysis,
was calculated for each gene of interest using the we screened cell-free CSF from a patient with MB and
ΔΔCT method, where cycle threshold values were compared the results to controls (CSF from two different
normalized to the level of cel-miR-39-3p (4464066, leukemia patients with no cerebral manifestation or
Journal of Cancer Metastasis and Treatment ¦ Volume 1 ¦ Issue 2 ¦ July 15, 2015 ¦ 69