Page 31 - Read Online
P. 31
Udukala et al. J Cancer Metastasis Treat 2020;6:25 I http://dx.doi.org/10.20517/2394-4722.2020.45 Page 5 of 13
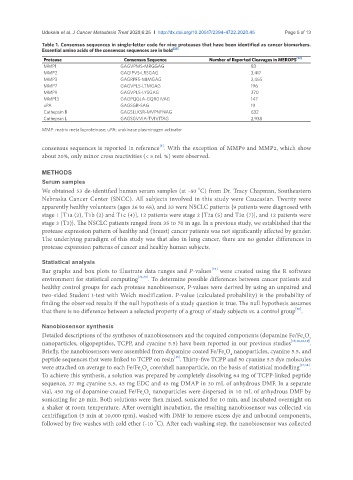
Table 1. Consensus sequences in single-letter code for nine proteases that have been identified as cancer biomarkers.
Essential amino acids of the consensus sequences are in bold [32]
Protease Consensus Sequence Number of Reported Cleavages in MEROPS [32]
MMP1 GAGVPMS-MRGGAG 83
MMP2 GAGIPVS-LRSGAG 3,417
MMP3 GAGRPFS-MIMGAG 2,465
MMP7 GAGVPLS-LTMGAG 196
MMP9 GAGVPLS-LYSGAG 370
MMP13 GAGPQGLA-GQRGIVAG 147
uPA GAGSGR-SAG 19
Cathepsin B GAGSLLKSR-MVPNFNAG 632
Cathepsin L GAGSGVVIA-TVIVITAG 2,938
MMP: matrix metalloproteinase; uPA: urokinase plasminogen activator
consensus sequences is reported in reference . With the exception of MMP9 and MMP2, which show
[5]
about 20%, only minor cross reactivities (< 5 rel. %) were observed.
METHODS
Serum samples
o
We obtained 53 de-identified human serum samples (at -80 C) from Dr. Tracy Chapman, Southeastern
Nebraska Cancer Center (SNCC). All subjects involved in this study were Caucasian. Twenty were
apparently healthy volunteers (ages 26 to 68), and 33 were NSCLC patients {9 patients were diagnosed with
stage 1 [T1a (3), T1b (2) and T1c (4)], 12 patients were stage 2 [T2a (5) and T3a (7)], and 12 patients were
stage 3 (T3)}. The NSCLC patients ranged from 35 to 70 in age. In a previous study, we established that the
protease expression pattern of healthy and (breast) cancer patients was not significantly affected by gender.
The underlying paradigm of this study was that also in lung cancer, there are no gender differences in
protease expression patterns of cancer and healthy human subjects.
Statistical analysis
[33]
Bar graphs and box plots to illustrate data ranges and P-values were created using the R software
environment for statistical computing [34,35] . To determine possible differences between cancer patients and
healthy control groups for each protease nanobiosensor, P-values were derived by using an unpaired and
two-sided Student t-test with Welch modification. P-value (calculated probability) is the probability of
finding the observed results if the null hypothesis of a study question is true. The null hypothesis assumes
[36]
that there is no difference between a selected property of a group of study subjects vs. a control group .
Nanobiosensor synthesis
Detailed descriptions of the syntheses of nanobiosensors and the required components (dopamine Fe/Fe O
4
3
nanoparticles, oligopeptides, TCPP, and cyanine 5.5) have been reported in our previous studies [15,16,18,19] .
Briefly, the nanobiosensors were assembled from dopamine coated Fe/Fe O nanoparticles, cyanine 5.5, and
4
3
[15]
peptide sequences that were linked to TCPP on resin . Thirty-five TCPP and 50 cyanine 5.5 dye molecules
were attached on average to each Fe/Fe O core/shell nanoparticle, on the basis of statistical modelling [37,38] .
4
3
To achieve this synthesis, a solution was prepared by completely dissolving 64 mg of TCPP-linked peptide
sequence, 37 mg cyanine 5.5, 45 mg EDC and 45 mg DMAP in 30 mL of anhydrous DMF. In a separate
vial, 450 mg of dopamine-coated Fe/Fe O nanoparticles were dispersed in 10 mL of anhydrous DMF by
3
4
sonicating for 20 min. Both solutions were then mixed, sonicated for 10 min, and incubated overnight on
a shaker at room temperature. After overnight incubation, the resulting nanobiosensor was collected via
centrifugation (5 min at 10,000 rpm), washed with DMF to remove excess dye and unbound components,
o
followed by five washes with cold ether (-10 C). After each washing step, the nanobiosensor was collected