Page 114 - Read Online
P. 114
Vidoni et al. J Cancer Metastasis Treat 2021;7:4 I http://dx.doi.org/10.20517/2394-4722.2020.95 Page 7 of 20
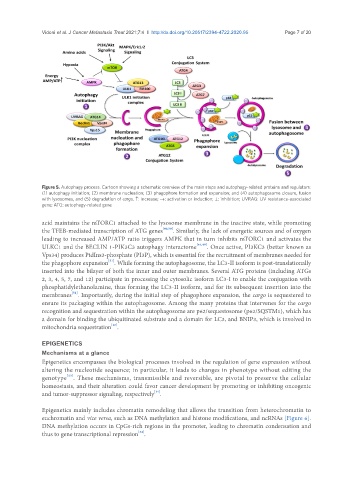
Figure 5. Autophagy process. Cartoon showing a schematic overview of the main steps and autophagy-related proteins and regulators:
(1) autophagy initiation; (2) membrane nucleation; (3) phagophore formation and expansion; and (4) autophagosome closure, fusion
with lysosomes, and (5) degradation of cargo. ↑: increase; →: activation or induction; ⊥: inhibition; UVRAG: UV resistance-associated
gene; ATG: autophagy-related gene
acid maintains the mTORC1 attached to the lysosome membrane in the inactive state, while promoting
the TFEB-mediated transcription of ATG genes [40,50] . Similarly, the lack of energetic sources and of oxygen
leading to increased AMP/ATP ratio triggers AMPK that in turn inhibits mTORC1 and activates the
ULKC1 and the BECLIN 1-PIK3C3 autophagy interactome [46,49] . Once active, PI3KC3 (better known as
Vps34) produces PtdIns3-phosphate (PI3P), which is essential for the recruitment of membranes needed for
[51]
the phagophore expansion . While forming the autophagosome, the LC3-II isoform is post-translationally
inserted into the bilayer of both the inner and outer membranes. Several ATG proteins (including ATGs
2, 3, 4, 5, 7, and 12) participate in processing the cytosolic isoform LC3-I to enable the conjugation with
phosphatidylethanolamine, thus forming the LC3-II isoform, and for its subsequent insertion into the
[52]
membranes . Importantly, during the initial step of phagophore expansion, the cargo is sequestered to
ensure its packaging within the autophagosome. Among the many proteins that intervenes for the cargo
recognition and sequestration within the autophagosome are p62/sequestosome (p62/SQSTM1), which has
a domain for binding the ubiquitinated substrate and a domain for LC3, and BNIP3, which is involved in
[46]
mitochondria sequestration .
EPIGENETICS
Mechanisms at a glance
Epigenetics encompasses the biological processes involved in the regulation of gene expression without
altering the nucleotide sequence; in particular, it leads to changes in phenotype without editing the
[53]
genotype . These mechanisms, transmissible and reversible, are pivotal to preserve the cellular
homeostasis, and their alteration could favor cancer development by promoting or inhibiting oncogenic
and tumor-suppressor signaling, respectively .
[14]
Epigenetics mainly includes chromatin remodeling that allows the transition from heterochromatin to
euchromatin and vice versa, such as DNA methylation and histone modifications, and ncRNAs [Figure 6].
DNA methylation occurs in CpGs-rich regions in the promoter, leading to chromatin condensation and
[54]
thus to gene transcriptional repression .