Page 93 - Read Online
P. 93
Page 419 Gutierrez et al. Cancer Drug Resist 2021;4:414-23 I http://dx.doi.org/10.20517/cdr.2020.113
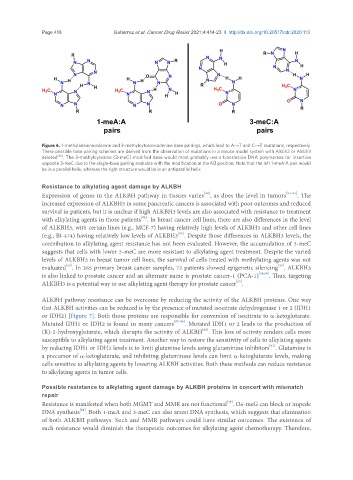
Figure 6. 1-methyladenine:adenine and 3-methylcytosine:adenine base pairings, which lead to A→T and C→T mutations, respectively.
These possible base pairing schemes are derived from the observation of mutations in a mouse model system with Alkbh2 or Alkbh3
deleted [48] . The 3-methylcytosine (3-meC) modified base would most probably use a translesion DNA polymerase for insertion
opposite 3-meC due to the single-base pairing available with the modification at the N3 position. Note that the left 1-meA:A pair would
be in a parallel helix, whereas the right structure would be in an antiparallel helix.
Resistance to alkylating agent damage by ALKBH
[50]
Expression of genes in the ALKBH pathway in tissues varies , as does the level in tumors [51-54] . The
increased expression of ALKBH3 in some pancreatic cancers is associated with poor outcomes and reduced
survival in patients, but it is unclear if high ALKBH3 levels are also associated with resistance to treatment
[52]
with alkylating agents in those patients . In breast cancer cell lines, there are also differences in the level
of ALKBH3, with certain lines (e.g., MCF-7) having relatively high levels of ALKBH3 and other cell lines
[53]
(e.g., Bt-474) having relatively low levels of ALKBH3 . Despite those differences in ALKBH3 levels, the
contribution to alkylating agent resistance has not been evaluated. However, the accumulation of 3-meC
suggests that cells with lower 3-meC are more resistant to alkylating agent treatment. Despite the varied
levels of ALKBH3 in breast tumor cell lines, the survival of cells treated with methylating agents was not
[53]
[53]
evaluated . In 265 primary breast cancer samples, 72 patients showed epigenetic silencing . ALKBH3
is also linked to prostate cancer and an alternate name is prostate cancer-1 (PCA-1) [55,56] . Thus, targeting
[55]
ALKBH3 is a potential way to use alkylating agent therapy for prostate cancer .
ALKBH pathway resistance can be overcome by reducing the activity of the ALKBH proteins. One way
that ALKBH activities can be reduced is by the presence of mutated isocitrate dehydrogenase 1 or 2 (IDH1
or IDH2) [Figure 7]. Both those proteins are responsible for conversion of isocitrate to α-ketoglutarate.
Mutated IDH1 or IDH2 is found in many cancers [57-59] . Mutated IDH1 or 2 leads to the production of
[60]
(R)-2-hydroxyglutarate, which disrupts the activity of ALKBH . This loss of activity renders cells more
susceptible to alkylating agent treatment. Another way to restore the sensitivity of cells to alkylating agents
[61]
by reducing IDH1 or IDH2 levels is to limit glutamine levels using glutaminase inhibitors . Glutamine is
a precursor of α-ketoglutarate, and inhibiting glutaminase levels can limit α-ketoglutarate levels, making
cells sensitive to alkylating agents by lowering ALKBH activities. Both these methods can reduce resistance
to alkylating agents in tumor cells.
Possible resistance to alkylating agent damage by ALKBH proteins in concert with mismatch
repair
Resistance is manifested when both MGMT and MMR are not functional . O6-meG can block or impede
[39]
[62]
DNA synthesis . Both 1-meA and 3-meC can also arrest DNA synthesis, which suggests that elimination
of both ALKBH pathways. Such and MMR pathways could have similar outcomes. The existence of
such resistance would diminish the therapeutic outcomes for alkylating agent chemotherapy. Therefore,