Page 89 - Read Online
P. 89
Page 415 Gutierrez et al. Cancer Drug Resist 2021;4:414-23 I http://dx.doi.org/10.20517/cdr.2020.113
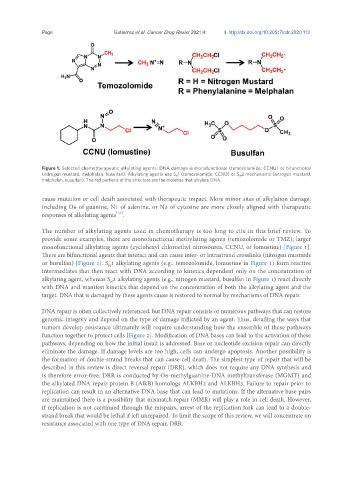
Figure 1. Selected chemotherapeutic alkylating agents. DNA damage is monofunctional (temozolomide, CCNU) or bifunctional
(nitrogen mustard, melphalan, busulfan). Alkylating agents use S N 1 (temozolomide, CCNU) or S N 2 mechanisms (nitrogen mustard,
melphalan, busulfan). The red portions of the structure are the moieties that alkylate DNA.
cause mutation or cell death associated with therapeutic impact. More minor sites of alkylation damage,
including O6 of guanine, N1 of adenine, or N3 of cytosine are more closely aligned with therapeutic
[1,2]
responses of alkylating agents .
The number of alkylating agents used in chemotherapy is too long to cite in this brief review. To
provide some examples, there are monofunctional methylating agents (temozolomide or TMZ), larger
monofunctional alkylating agents (cyclohexyl chloroethyl nitrosourea, CCNU, or lomustine) [Figure 1].
There are bifunctional agents that interact and can cause inter- or intrastrand crosslinks (nitrogen mustards
or busulfan) [Figure 1]. S 1 alkylating agents (e.g., temozolomide, lomustine in Figure 1) form reactive
N
intermediates that then react with DNA according to kinetics dependent only on the concentration of
alkylating agent, whereas S 2 alkylating agents (e.g., nitrogen mustard, busulfan in Figure 1) react directly
N
with DNA and manifest kinetics that depend on the concentration of both the alkylating agent and the
target. DNA that is damaged by these agents cause is restored to normal by mechanisms of DNA repair.
DNA repair is often collectively referenced, but DNA repair consists of numerous pathways that can restore
genomic integrity and depend on the type of damage inflicted by an agent. Thus, detailing the ways that
tumors develop resistance ultimately will require understanding how the ensemble of these pathways
function together to protect cells [Figure 2]. Modification of DNA bases can lead to the activation of these
pathways, depending on how the initial insult is addressed. Base or nucleotide excision repair can directly
eliminate the damage. If damage levels are too high, cells can undergo apoptosis. Another possibility is
the formation of double-strand breaks that can cause cell death. The simplest type of repair that will be
described in this review is direct reversal repair (DRR), which does not require any DNA synthesis and
is therefore error-free. DRR is conducted by O6-methylguanine-DNA methyltransferase (MGMT) and
the alkylated DNA repair protein B (AlkB) homologs ALKBH2 and ALKBH3. Failure to repair prior to
replication can result in an alternative DNA base that can lead to mutations. If the alternative base pairs
are maintained there is a possibility that mismatch repair (MMR) will play a role in cell death. However,
if replication is not continued through the mispairs, arrest of the replication fork can lead to a double-
strand break that would be lethal if left unrepaired. To limit the scope of this review, we will concentrate on
resistance associated with one type of DNA repair, DRR.