Page 201 - Read Online
P. 201
Alonso-Peña et al. Cancer Drug Resist 2019;2:680-709 I http://dx.doi.org/10.20517/cdr.2019.006 Page 697
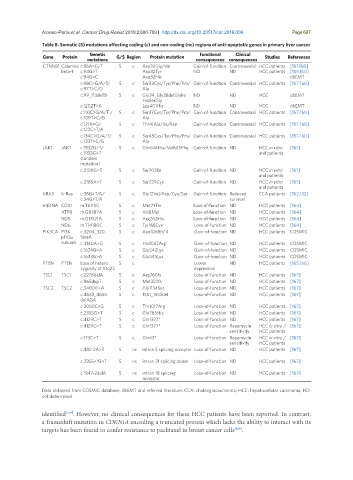
Table 8. Somatic (S) mutations affecting coding (c) and non-coding (nc) regions of anti-apoptotic genes in primary liver cancer
Genetic Functional Clinical
Gene Protein G/S Region Protein mutation Studies References
mutations consequences consequences
CTNNB1 Catenine c.95A>G/T S c Asp32Gly/Val Gain-of-function Controversial HCC patients [157,158]
beta-1 c.94G>T Asp32Tyr ND ND HCC patients [159,160]
c.94G>C Asp32His dbEMT
c.98C>G/A/T/ S c Ser33Cys/Tyr/Phe/Pro/ Gain-of-function Controversial HCC patients [157-160]
c.97T>C/G Ala
c.99_113del15 S c Gly34_Gly38delGlyIle ND ND HCC dbEMT
HisSerGly
c.1202T>A Leu401His ND ND HCC dbEMT
c.110C>G/A/T / S c Ser37Cys/Tyr/Phe/Pro/ Gain-of-function Controversial HCC patients [157-160]
c.109T>C/G Ala
c.121A>G/ S c Thr41Ala/Ile/Asn Gain-of-function Controversial HCC patients [157-160]
c.122C>T/A
c.134C>G/A/T/ S c Ser45Cys/Tyr/Phe/Pro/ Gain-of-function Controversial HCC patients [157-160]
c.133T>C/G Ala
JAK1 JAK1 c.1932G>T/ S c Gln644His/Val645Phe Gain-of-function ND HCC in vitro [161]
c.1933G>T and patients
(tandem
mutation)
c.2108G>T S c Ser703Ile Gain-of-function ND HCC in vitro [161]
and patients
c.2185A>T S c Ser729Cys Gain-of-function ND HCC in vitro [161]
and patients
KRAS K-Ras c.35G>T/A/ S c Gly12Val/Asp/Cys/Ser Gain-of-function Reduced CCA patients [162,132]
c.34G>T/A survival
mtDNA COX1 m.T6115C S c Met71Thr Loss-of-function ND HCC patients [164]
ATP8 m.G8387A S c Val8Met Loss-of-function ND HCC patients [164]
ND5 m.G13121A S c Arg262His Loss-of-function ND HCC patients [164]
ND6 m.T14180C S c Tyr165Cys Loss-of-function ND HCC patients [164]
PIK3CA PI3K c.3204_320 S c Asn1068fs*4 Gain-of-function ND HCC patients COSMIC
p110α 5insA
subunit c.3140A>G S c His1047Arg Gain-of-function ND HCC patients COSMIC
c.1624G>A S c Glu542Lys Gain-of-function ND HCC patients COSMIC
c.1633G>A S c Glu545Lys Gain-of-function ND HCC patients COSMIC
PTEN PTEN Loss of hetero S c Lower ND HCC patients [165,166]
zygosity at 10q23 expression
TSC1 TSC1 c.2278delA S c Arg760fs Loss-of-function ND HCC patients [167]
c.965dupT S c Met322fs Loss-of-function ND HCC patients [167]
TSC2 TSC2 c.3400G>A S c Gly1134Ser Loss-of-function ND HCC patients [167]
c.4653_4655 S c 1551_1552del Loss-of-function ND HCC patients [167]
delAGA
c.3050C>G S c Thr1017Arg Loss-of-function ND HCC patients [167]
c.2355G>T S c Gln785His Loss-of-function ND HCC patients [167]
c.4129C>T S c Gln1377* Loss-of-function ND HCC patients [167]
c.4129C>T S c Gln1377* Loss-of-function Rapamycin HCC in vitro / [167]
sensitivity HCC patients
c.173C>T S c Gln63* Loss-of-function Rapamycin HCC in vitro / [167]
sensitivity HCC patients
c.482-2A>T S nc intron 5 splicing acceptor Loss-of-function ND HCC patients [167]
c.2355+1G>T S nc intron 21 splicing donor Loss-of-function ND HCC patients [167]
c.1947-2delA S nc intron 18 splicing Loss-of-function ND HCC patients [167]
acceptor
Data obtained from COSMIC database, dbEMT and referred literature. CCA: cholangiocarcinoma; HCC: hepatocellular carcinoma; ND:
not determined
identified . However, no clinical consequences for these HCC patients have been reported. In contrast,
[119]
a frameshift mutation in CDKN1A encoding a truncated protein which lacks the ability to interact with its
targets has been found to confer resistance to paclitaxel in breast cancer cells .
[168]