Page 193 - Read Online
P. 193
Alonso-Peña et al. Cancer Drug Resist 2019;2:680-709 I http://dx.doi.org/10.20517/cdr.2019.006 Page 689
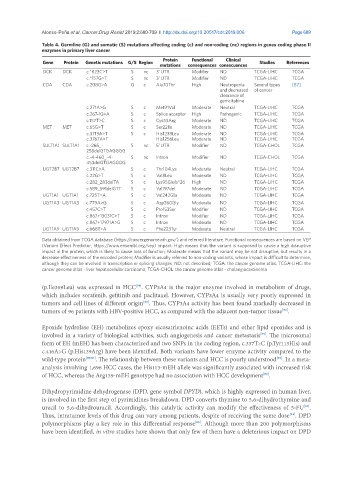
Table 4. Germline (G) and somatic (S) mutations affecting coding (c) and non-coding (nc) regions in genes coding phase II
enzymes in primary liver cancer
Protein Functional Clinical
Gene Protein Genetic mutations G/S Region Studies References
mutations consequences consecuences
DCK DCK c.*823C>T S nc 3’ UTR Modifier ND TCGA-LIHC TCGA
c.*157G>T S nc 3’ UTR Modifier ND TCGA-LIHC TCGA
CDA CDA c.208G>A G c Ala70Thr High Neutropenia Several types [87]
and decreased of cancer
clearance of
gemcitabine
c.271A>G S c Met91Val Moderate Neutral TCGA-LIHC TCGA
c.267-1G>A S c Splice acceptor High Pathogenic TCGA-LIHC TCGA
c.157T>C S c Cys53Arg Moderate ND TCGA-LIHC TCGA
MET MET c.65G>T S c Ser22Ile Moderate ND TCGA-LIHC TCGA
c.3713A>T S c His1238Leu Moderate ND TCGA-LIHC TCGA
c.3767A>T His1256Leu Moderate ND TCGA-LIHC TCGA
SULT1A1 SULT1A1 c.-265_- S nc 5’ UTR Modifier ND TCGA-CHOL TCGA
258delGTGAGGGG
c.-4-460_-4- S nc Intron Modifier ND TCGA-CHOL TCGA
453delGTGAGGGG
UGT2B7 UGT2B7 c.311C>A S c Thr104Lys Moderate Neutral TCGA-LIHC TCGA
c.22G>T S c Val8Leu Moderate ND TCGA-LIHC TCGA
c.282_283delTA S c Lys95Glufs*26 High ND TCGA-LIHC TCGA
c.589_591delGTT S c Val197del Moderate ND TCGA-LIHC TCGA
UGT1A1 UGT1A1 c.725T>A S c Val242Glu Moderate ND TCGA-LIHC TCGA
UGT1A3 UGT1A3 c.779A>G S c Asp260Gly Moderate ND TCGA-LIHC TCGA
c.457C>T S c Pro153Ser Modifier ND TCGA-LIHC TCGA
c.867+13031C>T S c Intron Modifier ND TCGA-LIHC TCGA
c.867+17971A>G S c Intron Moderate ND TCGA-LIHC TCGA
UGT1A9 UGT1A9 c.668T>A S c Phe223Tyr Moderate Neutral TCGA-LIHC TCGA
Data obtained from TCGA database (https://cancergenome.nih.gov/) and referred literature. Functional consequences are based on VEP
(Variant Effect Predictor; https://www.ensembl.org/vep) impact: High means that the variant is supposed to cause a high disruptive
impact in the protein, which is likely to cause loss of function; Moderate means that the variant may be not disruptive, but results in a
decrease effectiveness of the encoded protein; Modifier is usually referred to non-coding variants, whose impact is difficult to determine,
although they can be involved in transcription or splicing changes. ND: not described; TCGA: the cancer genome atlas; TCGA-LIHC: the
cancer genome atlas - liver hepatocellular carcinoma; TCGA-CHOL: the cancer genome atlas - cholangiocarcinoma
(p.Ile359Leu) was expressed in HCC . CYP3A4 is the major enzyme involved in metabolism of drugs,
[79]
which includes sorafenib, gefitinib and paclitaxel. However, CYP3A4 is usually very poorly expressed in
tumors and cell lines of different origin . Thus, CYP3A4 activity has been found markedly decreased in
[88]
tumors of 96 patients with HBV-positive HCC, as compared with the adjacent non-tumor tissue .
[85]
Epoxide hydrolase (EH) metabolizes epoxy eicosatrienoinc acids (EETs) and other lipid epoxides and is
involved in a variety of biological activities, such angiogenesis and cancer metastasis . The microsomal
[89]
form of EH (mEH) has been characterized and two SNPs in the coding region, c.337T>C (p.Tyr113His) and
c.416A>G (p.His139Arg) have been identified. Both variants have lower enzyme activity compared to the
wild-type protein [90,91] . The relationship between these variants and HCC is poorly understood . In a meta-
[92]
analysis involving 1,696 HCC cases, the His113-mEH allele was significantly associated with increased risk
of HCC, whereas the Arg139-mEH genotype had no association with HCC development .
[80]
Dihydropyrimidine dehydrogenase (DPD, gene symbol DPYD), which is highly expressed in human liver,
is involved in the first step of pyrimidines breakdown. DPD converts thymine to 5,6-dihydrothymine and
uracil to 5,6-dihydrouracil. Accordingly, this catalytic activity can modify the effectiveness of 5-FU .
[93]
Thus, intratumor levels of this drug can vary among patients, despite of receiving the same dose . DPD
[94]
polymorphisms play a key role in this differential response . Although more than 200 polymorphisms
[86]
have been identified, in vitro studies have shown that only few of them have a deleterious impact on DPD