Page 192 - Read Online
P. 192
Page 688 Alonso-Peña et al. Cancer Drug Resist 2019;2:680-709 I http://dx.doi.org/10.20517/cdr.2019.006
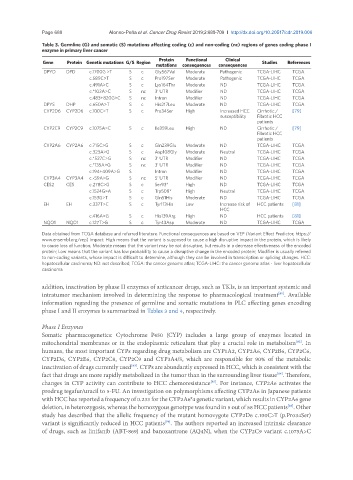
Table 3. Germline (G) and somatic (S) mutations affecting coding (c) and non-coding (nc) regions of genes coding phase I
enzyme in primary liver cancer
Protein Functional Clinical
Gene Protein Genetic mutations G/S Region Studies References
mutations consequences consequences
DPYD DPD c.1700G >T S c Gly567Val Moderate Pathogenic TCGA-LIHC TCGA
c.589C>T S c Pro197Ser Moderate Pathogenic TCGA-LIHC TCGA
c.491A>C S c Lys164Thr Moderate ND TCGA-LIHC TCGA
c.*102A>C S nc 3’ UTR Modifier ND TCGA-LIHC TCGA
c.483+820G>C S nc Intron Modifier ND TCGA-LIHC TCGA
DPYS DHP c.650A>T S c His217Leu Moderate ND TCGA-LIHC TCGA
CYP2D6 CYP2D6 c.100C>T S c Pro34Ser High Increased HCC Cirrhotic / [79]
susceptibility Fibrotic HCC
patients
CYP2C9 CYP2C9 c.1075A>C S c Ile359Leu High ND Cirrhotic / [79]
Fibrotic HCC
patients
CYP2A6 CYP2A6 c.715C>G S c Gln239Glu Moderate ND TCGA-LIHC TCGA
c.323A>G S c Asp108Gly Moderate Neutral TCGA-LIHC TCGA
c.*527C>G S nc 3’ UTR Modifier ND TCGA-LIHC TCGA
c.*135A>G S nc 3’ UTR Modifier ND TCGA-LIHC TCGA
c.194+409A>G S Intron Modifier ND TCGA-LIHC TCGA
CYP3A4 CYP3A4 c.-59A>G S nc 5‘ UTR Modifier ND TCGA-LIHC TCGA
CES2 CES c.278C>G S c Ser93* High ND TCGA-LIHC TCGA
c.1524G>A S c Trp508* High Neutral TCGA-LIHC TCGA
c.153G>T S c Gln51His Moderate ND TCGA-LIHC TCGA
EH EH c.337T>C S c Tyr113His Low Increase risk of HCC patients [81]
HCC
c.416A>G S c His139Arg High ND HCC patients [81]
NQO1 NQO1 c.127T>G S c Tyr43Asp Moderate ND TCGA-LIHC TCGA
Data obtained from TCGA database and referred literature. Functional consequences are based on VEP (Variant Effect Predictor; https://
www.ensembl.org/vep) impact: High means that the variant is supposed to cause a high disruptive impact in the protein, which is likely
to cause loss of function; Moderate means that the variant may be not disruptive, but results in a decrease effectiveness of the encoded
protein; Low means that the variant has low probability to cause a disruptive change in the encoded protein; Modifier is usually referred
to non-coding variants, whose impact is difficult to determine, although they can be involved in transcription or splicing changes. HCC:
hepatocellular carcinoma; ND: not described; TCGA: the cancer genome atlas; TCGA-LIHC: the cancer genome atlas - liver hepatocellular
carcinoma
addition, inactivation by phase II enzymes of anticancer drugs, such as TKIs, is an important systemic and
intratumor mechanism involved in determining the response to pharmacological treatment . Available
[80]
information regarding the presence of germline and somatic mutations in PLC affecting genes encoding
phase I and II enzymes is summarized in Tables 3 and 4, respectively.
Phase I Enzymes
Somatic pharmacogenetics: Cytochrome P450 (CYP) includes a large group of enzymes located in
[82]
mitochondrial membranes or in the endoplasmic reticulum that play a crucial role in metabolism . In
humans, the most important CYPs regarding drug metabolism are CYP1A2, CYP2A6, CYP2B6, CYP2C6,
CYP2D6, CYP2E6, CYP2C8, CYP2C9 and CYP3A4/5, which are responsible for 90% of the metabolic
inactivation of drugs currently used . CYPs are abundantly expressed in HCC, which is consistent with the
[83]
[84]
fact that drugs are more rapidly metabolized in the tumor than in the surrounding liver tissue . Therefore,
changes in CYP activity can contribute to HCC chemoresistance . For instance, CYP2A6 activates the
[85]
prodrug tegafur/uracil to 5-FU. An investigation on polymorphisms affecting CYP2A6 in Japanese patients
with HCC has reported a frequency of 0.233 for the CYP2A6*4 genetic variant, which results in CYP2A6 gene
deletion, in heterozygosis, whereas the homozygous genotype was found in 5 out of 58 HCC patients . Other
[86]
study has described that the allelic frequency of the mutant homozygote CYP2D6 c.100C>T (p.Pro34Ser)
[79]
variant is significantly reduced in HCC patients . The authors reported an increased intrinsic clearance
of drugs, such as linifanib (ABT-869) and banoxantrone (AQ4N), when the CYP2C9 variant c.1075A>C