Page 396 - Read Online
P. 396
Page 10 of 13 Iqbal et al. Vessel Plus 2019;3:40 I http://dx.doi.org/10.20517/2574-1209.2019.28
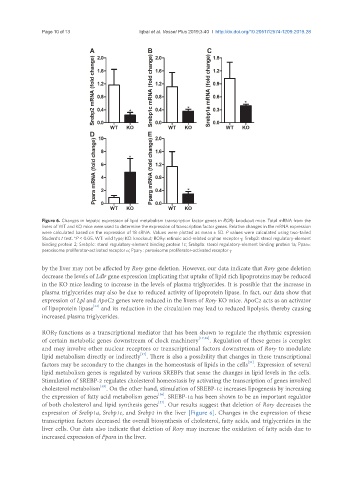
Figure 6. Changes in hepatic expression of lipid metabolism transcription factor genes in RORγ knockout mice. Total mRNA from the
livers of WT and KO mice were used to determine the expression of transcription factor genes. Relative changes in the mRNA expression
were calculated based on the expression of 18 sRNA. Values were plotted as mean ± SD. P values were calculated using two-tailed
Student’s t test. *P < 0.05. WT: wild type; KO: knockout; RORγ: retinoic acid-related orphan receptor γ; Srebp2: sterol regulatory-element
binding protein 2; Srebp1c: sterol regulatory-element binding protein 1c; Srebp1a: sterol regulatory-element binding protein 1a; Pparα:
peroxisome proliferator-activated receptor α; Pparγ : peroxisome proliferator-activated receptor γ
by the liver may not be affected by Rorγ gene deletion. However, our data indicate that Rorγ gene deletion
decrease the levels of Ldlr gene expression implicating that uptake of lipid rich lipoproteins may be reduced
in the KO mice leading to increase in the levels of plasma triglycerides. It is possible that the increase in
plasma triglycerides may also be due to reduced activity of lipoprotein lipase. In fact, our data show that
expression of Lpl and ApoC2 genes were reduced in the livers of Rorγ KO mice. ApoC2 acts as an activator
[33]
of lipoprotein lipase and its reduction in the circulation may lead to reduced lipolysis, thereby causing
increased plasma triglycerides.
RORγ functions as a transcriptional mediator that has been shown to regulate the rhythmic expression
of certain metabolic genes downstream of clock machinery [17,18] . Regulation of these genes is complex
and may involve other nuclear receptors or transcriptional factors downstream of Rorγ to modulate
[17]
lipid metabolism directly or indirectly . There is also a possibility that changes in these transcriptional
[34]
factors may be secondary to the changes in the homeostasis of lipids in the cells . Expression of several
lipid metabolism genes is regulated by various SREBPs that sense the changes in lipid levels in the cells.
Stimulation of SREBP-2 regulates cholesterol homeostasis by activating the transcription of genes involved
[35]
cholesterol metabolism . On the other hand, stimulation of SREBP-1c increases lipogenesis by increasing
[36]
the expression of fatty acid metabolism genes . SREBP-1a has been shown to be an important regulator
of both cholesterol and lipid synthesis genes . Our results suggest that deletion of Rorγ decreases the
[37]
expression of Srebp1a, Srebp1c, and Srebp2 in the liver [Figure 6]. Changes in the expression of these
transcription factors decreased the overall biosynthesis of cholesterol, fatty acids, and triglycerides in the
liver cells. Our data also indicate that deletion of Rorγ may increase the oxidation of fatty acids due to
increased expression of Ppara in the liver.