Page 93 - Read Online
P. 93
Page 8 of 15 Longhi et al. Microbiome Res Rep 2024;3:4 https://dx.doi.org/10.20517/mrr.2023.02
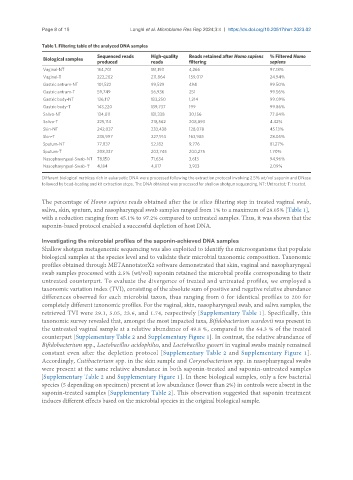
Table 1. Filtering table of the analyzed DNA samples
Sequenced reads High-quality Reads retained after Homo sapiens % Filtered Homo
Biological samples
produced reads filtering sapiens
Vaginal-NT 154,701 151,193 4,266 97.18%
Vaginal-T 222,202 211,864 159,017 24.94%
Gastric antrum-NT 101,522 99,529 494 99.50%
Gastric antrum-T 59,749 56,936 251 99.56%
Gastric body-NT 136,117 133,250 1,214 99.09%
Gastric body-T 143,220 139,737 199 99.86%
Saliva-NT 134,011 131,338 30,156 77.04%
Saliva-T 225,114 218,562 208,893 4.42%
Skin-NT 242,037 233,438 128,078 45.13%
Skin-T 236,997 227,914 163,985 28.05%
Sputum-NT 77,837 52,182 9,776 81.27%
Sputum-T 208,337 203,745 200,275 1.70%
Nasopharyngeal-Swab-NT 78,150 71,634 3,613 94.96%
Nasopharyngeal-Swab-T 4,184 4,017 3,933 2.09%
Different biological matrices rich in eukaryotic DNA were processed following the extraction protocol involving 2.5% wt/vol saponin and DNase
followed by bead-beating and kit extraction steps. The DNA obtained was processed for shallow shotgun sequencing. NT: Untreated; T: treated.
The percentage of Homo sapiens reads obtained after the in silico filtering step in treated vaginal swab,
saliva, skin, sputum, and nasopharyngeal swab samples ranged from 1% to a maximum of 28.05% [Table 1],
with a reduction ranging from 45.1% to 97.2% compared to untreated samples. Thus, it was shown that the
saponin-based protocol enabled a successful depletion of host DNA.
Investigating the microbial profiles of the saponin-achieved DNA samples
Shallow shotgun metagenomic sequencing was also exploited to identify the microorganisms that populate
biological samples at the species level and to validate their microbial taxonomic composition. Taxonomic
profiles obtained through METAnnotatorX2 software demonstrated that skin, vaginal and nasopharyngeal
swab samples processed with 2.5% (wt/vol) saponin retained the microbial profile corresponding to their
untreated counterpart. To evaluate the divergence of treated and untreated profiles, we employed a
taxonomic variation index (TVI), consisting of the absolute sum of positive and negative relative abundance
differences observed for each microbial taxon, thus ranging from 0 for identical profiles to 200 for
completely different taxonomic profiles. For the vaginal, skin, nasopharyngeal swab, and saliva samples, the
retrieved TVI were 29.1, 5.05, 23.6, and 1.74, respectively [Supplementary Table 1]. Specifically, this
taxonomic survey revealed that, amongst the most impacted taxa, Bifidobacterium scardovii was present in
the untreated vaginal sample at a relative abundance of 49.8 %, compared to the 64.3 % of the treated
counterpart [Supplementary Table 2 and Supplementary Figure 1]. In contrast, the relative abundance of
Bifidobacterium spp., Lactobacillus acidophilus, and Lactobacillus gasseri in vaginal swabs mainly remained
constant even after the depletion protocol [Supplementary Table 2 and Supplementary Figure 1].
Accordingly, Cutibacterium spp. in the skin sample and Corynebacterium spp. in nasopharyngeal swabs
were present at the same relative abundance in both saponin-treated and saponin-untreated samples
[Supplementary Table 2 and Supplementary Figure 1]. In these biological samples, only a few bacterial
species (5 depending on specimen) present at low abundance (lower than 2%) in controls were absent in the
saponin-treated samples [Supplementary Table 2]. This observation suggested that saponin treatment
induces different effects based on the microbial species in the original biological sample.