Page 24 - Read Online
P. 24
Lugli et al. Microbiome Res Rep 2023;2:15 https://dx.doi.org/10.20517/mrr.2022.21 Page 11 of 16
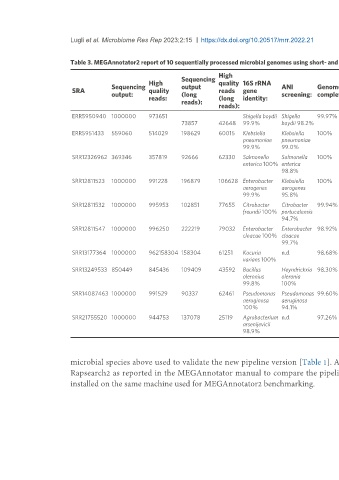
Table 3. MEGAnnotator2 report of 10 sequentially processed microbial genomes using short- and long-read technologies
High
Sequencing Average
High quality 16S rRNA Number Number Number Number
Sequencing output ANI Genome Genome Average coverage Genome
SRA quality reads gene of of of rRNA of tRNA
output: (long screening: completeness: contamination: coverage: (long length:
reads: (long identity: contigs: genes: genes: genes:
reads): reads):
reads):
ERR5950940 1000000 973651 Shigella boydii Shigella 99.97% 0.75% 75 39 55 6,606,912 6,860 22 97
73857 42648 99.9% boydii 98.2%
ERR5951433 559060 514029 198629 60015 Klebsiella Klebsiella 100% 1.04% 45 86 4 5,595,850 5,217 25 88
pneumoniae pneumoniae
99.9% 99.0%
SRR12326962 369346 357819 92666 62330 Salmonella Salmonella 100% 0.78% 32 97 5 5,310,747 5,134 22 92
enterica 100% enterica
98.8%
SRR12811523 1000000 991228 196879 106628 Enterobacter Klebsiella 100% 1.11% 49 99 8 5,567,807 5,281 25 90
aerogenes aerogenes
99.9% 95.8%
SRR12811532 1000000 995953 102851 77655 Citrobacter Citrobacter 99.94% 1.86% 48 99 16 5,693,281 5,598 25 86
freundii 100% portucalensis
94.7%
SRR12811547 1000000 996250 222219 79032 Enterobacter Enterobacter 98.92% 1.06% 50 98 19 5,496,621 5,248 25 87
cloacae 100% cloacae
99.7%
SRR13177364 1000000 962158304 158304 61251 Kocuria n.d. 98.68% 0% 163 120 2 2,848,142 2,432 9 48
varians 100%
SRR13249533 850449 845436 109409 43592 Bacillus Heyndrickxia 98.30% 6.27% 76 68 2 5,364,016 5,378 36 145
oleronius oleronia
99.8% 100%
SRR14087463 1000000 991529 90337 62461 Pseudomonas Pseudomonas 99.60% 0.17% 41 66 14 6,729,107 6,213 12 68
aeruginosa aeruginosa
100% 94.1%
SRR21755520 1000000 944753 137078 25119 Agrobacterium n.d. 97.26% 3.31% 46 52 4 5,773,279 5,401 15 60
arsenijevicii
98.9%
microbial species above used to validate the new pipeline version [Table 1]. An updated RefSeq database was downloaded from NCBI and formatted using
Rapsearch2 as reported in the MEGAnnotator manual to compare the pipeline efficiency. Furthermore, the last version of the MIRA assembler has been
installed on the same machine used for MEGAnnotator2 benchmarking.